Figure 4.
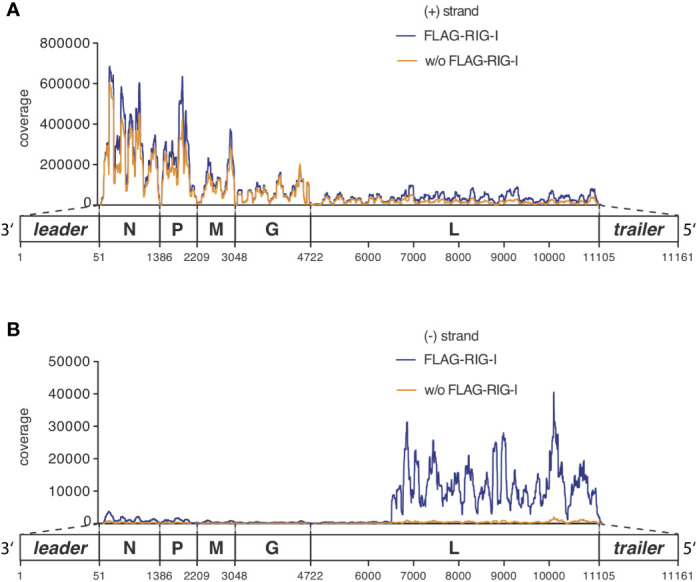
RNAs co-immunoprecipitated with anti-RIG-I-coated sepharose beads from lysates of VSV-infected cells consist mostly of unspecifically bound viral and endogenous mRNA transcripts but are specifically enriched for viral genomic sequences of negative orientation. HEK 293 cells (40 x 106) expressing either FLAG-RIG-I or not were infected with VSV (MOI=1). 9 hours later lysates were prepared and RIG-I/RNA complexes were immunoprecipitated with anti-FLAG antibody-coupled sepharose beads. After elution of the protein/RNA complexes from the beads, RNA was purified from the eluate and used to generate cDNA libraries for next generation sequencing on an Illumina genome analyzer. The read sequences were aligned to the VSV genome reference in (A) sense or (B) antisense orientation and are depicted on the y-axis as coverage meaning the number of sequence reads that cover each specific position of the reference genome represented on the x-axis. The x-axis has a resolution down to single nucleotide positions. A schematic representation of the VSV genome underlines the x-axis and is in areas of the trailer and leader sequences not true to scale. One representative experiment out of two is shown.
