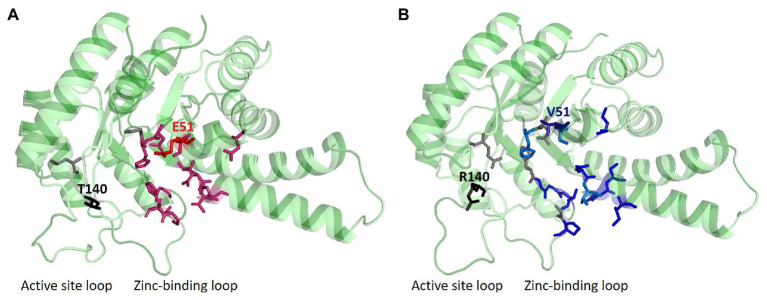
Figure 2.
Ligand and structure prediction of FBAC and FBAP. (A) 3D model of the FBAC showing the protein structure, which was depicted with PyMOL (DeLano, 2002), and predicted ligands via COACH-D (Wu et al., 2018b) analysis. (B) 3D protein model of FBAP. In gray, residues that are important for the zinc-binding are shown; while in red residues that are necessary for binding the substrate FBP are plotted. Blue residues are representing important sites for GAP or DHAP interaction. Sites for SDM are marked in the 3D structures of the protein models (E51V, T140R). The 3D structure of the protein models indicates typical triosephosphate isomerase (TIM) barrels.