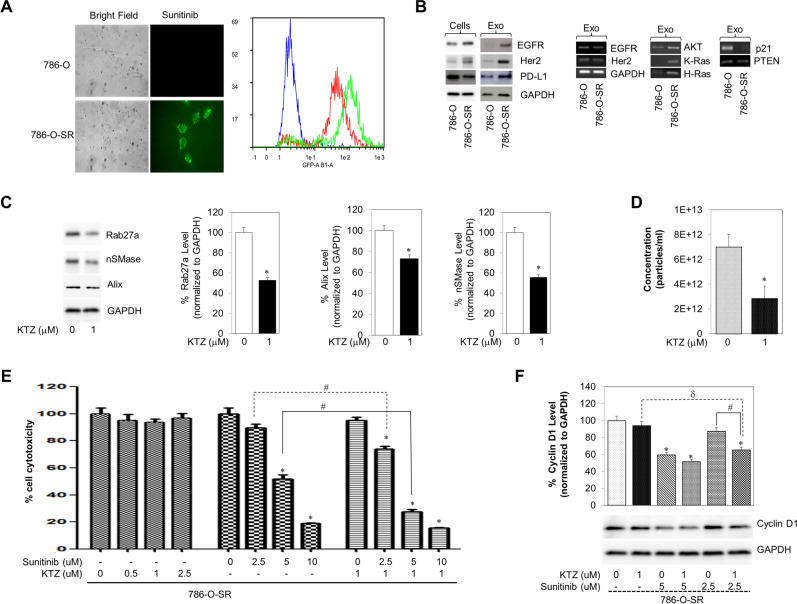
Figure 5.
Effect of KTZ co-exposure on 786-O or 786-O-SR cells. (A; left panel) Typical fluorescence microscope image of 786-O-SR cells following treatment with 5 µM sunitinib, exhibiting fluorescent granules (magnification, × 40). (A; right panel) Sunitinib autofluorescence in control (Blue) or sunitinib-treated 2.5 μM (Red), and 5 μM (Green) 786-O cells after incubation for 24 h by MACSQuant analysis. This figure was generated using MACSQuantify Software version 2.1.3. This software can be found at https://www.miltenyibiotec.com/US-en/products/macsquantify-software.html#130-094-556. (B-Left panel) Total cells and exosome protein lysates were prepared from 786-O and 786-O-SR cells. Proteins were subjected to immunoblot analysis with antibodies against EGFR, Her2, PD-L1, and GAPDH. (B-right panel) Conventional PCR analysis of EGFR, Her2, H-Ras, K-Ras and GAPDHs in the exosome derived from 786-O and 786-O-SR cells. (C) KTZ significantly inhibited the protein expression of Alix, nSMase and Rab27a in a dose-dependent manner in 786-O-SR. (D) qNano-IZON particle quantitative analysis (NP-100 nanopore) depicting a significant decrease in exosome concentrations (50–200 nm size) in the CM of 786-O-SR cells treated with KTZ compared to vehicle treated control. *Denotes significance at p < 0.05 compared to controls and was calculated using GraphPad Prism. (E) MTT cell proliferation assays showed that KTZ potentiated sunitinib inhibition of 786-O—Sunitinib Resistance (786-O-SR) cells growth. KTZ synergistically enhanced Sunitinib's cytotoxicity to 786-O-SR cells when the two drugs were combined at the IC50 concentration equivalent for each drug. (F) The 786-O-SR cells were treated with the indicated concentrations of sunitinib, ketoconazole or combination drugs for 48 h. Lysates were analyzed by western blot analysis using the indicated antibodies. Relative expression levels of cyclin D1 protein were compared between control cells and cells treated with the indicated drugs by western blot. GAPDH immunoblot used as loading control. Error bars represent SD. Three independent experiments were performed in triplicate. *Denotes significance at p < 0.05 compared to controls. dDenotes significance at p < 0.05 compared between KTZ-treated group and combination-treated group. Full blots are presented in Supplementary Fig. S6. #Denotes significance at p < 0.05 compared between sunitinib-treated group and combination-treated group and was calculated using GraphPad Prism. Data for EVs and exosomes was captured using IZON’s Control Suite software version 3.4.2.48, https://support.izon.com/how-can-i-get-the-latest-software-release.