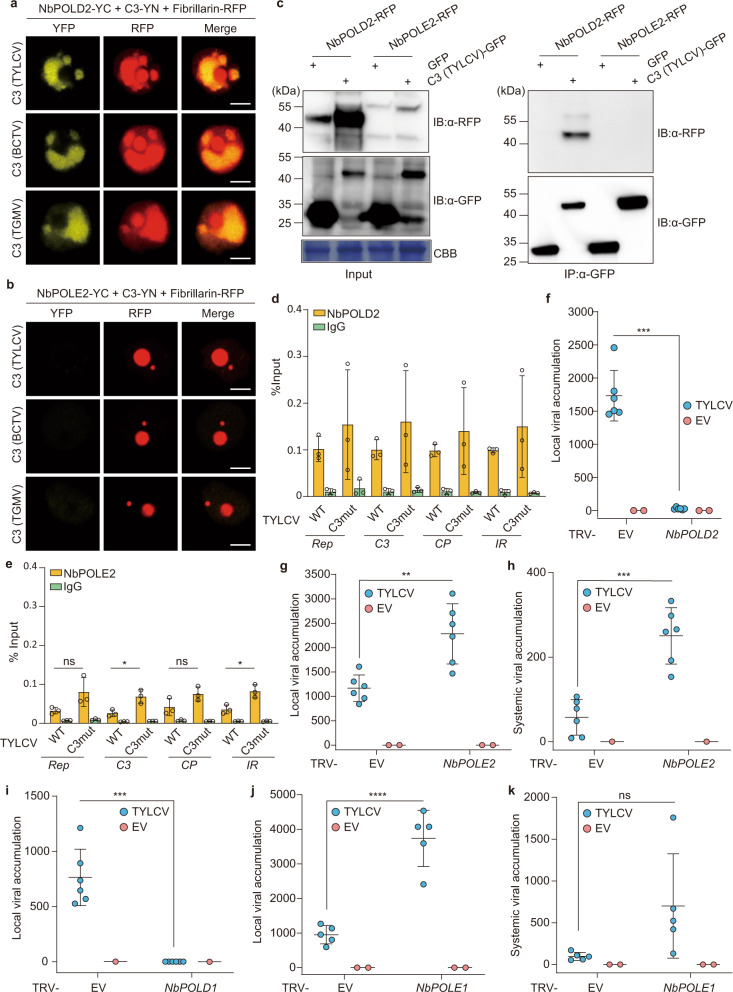
Fig. 2. DNA polymerase δ, but not DNA polymerase ε, interacts with the geminivirus-encoded C3 protein and is required for geminiviral replication.
a, b NbPOLD2 (a), but not NbPOLE2 (b), interacts with C3 from TYLCV, BCTV, and TGMV in BiFC assays upon transient expression in N. benthamiana. Fibrillarin-RFP marks the nucleolus and the Cajal body. Scale bar: 5 µm. Negative controls are shown in Supplementary Fig. 1. c C3-GFP co-immunoprecipitates NbPOLD2-RFP (left), but not NbPOLE2-RFP (right), upon transient expression in N. benthamiana. IP: immunoprecipitate; IB: immunoblotting; CBB: Coomassie brilliant blue. The predicted protein sizes are as follows: NbPOLD2-RFP, ~46.5 kDa; NbPOLE2-RFP, ~63 kDa; C3 (TYLCV)-GFP, ~42 kDa; GFP, ~26 kDa. Full blots and membranes can be found in the Source data file. d, e NbPOLD2 (d) and NbPOLE2 (e) bind the TYLCV genome in ChIP assays. The location of the amplified sequences at different genomic regions is shown in Supplementary Fig. 3a; results for additional genomic regions are shown in Supplementary Fig. 7. Data are the mean of three independent biological replicates; error bars indicate SD. Asterisks indicate a statistically significant difference according to a two-sided Student’s t test (*P < 0.05). ns: not significant. f, g Viral accumulation in local TYLCV infections (3 days post inoculation) in POLD2-silenced (TRV-NbPOLD2) (f), POLE2-silenced (TRV-NbPOLE2) (g), or control (TRV) N. benthamiana plants measured by qPCR. Plants inoculated with the empty vector (EV) are used as negative control. Data are the mean of six independent biological replicates; error bars represent SD. The phenotype of silenced plants and silencing efficiency are presented in Supplementary Fig. 4. h Viral accumulation in systemic TYLCV infections (2 weeks post inoculation) in POLE2-silenced (TRV-NbPOLE2) or control (TRV) N. benthamiana plants measured by qPCR. Plants inoculated with the empty vector (EV) are used as negative control. Data are the mean of six independent biological replicates; error bars represent SD. The 25S ribosomal DNA interspacer (ITS) was used as a reference gene; values are presented relative to ITS. i, j Viral accumulation in local TYLCV infections (3 days post inoculation) in POLD1-silenced (TRV-NbPOLD1) (i), POLE1-silenced (TRV-NbPOLE1) (j), or control (TRV) N. benthamiana plants measured by qPCR. Plants inoculated with the empty vector (EV) are used as negative control. Data are the mean of six (i) or five (j) independent biological replicates; error bars represent SD. The phenotype of silenced plants and silencing efficiency are presented in Supplementary Fig. 4. k Viral accumulation in systemic TYLCV infections (2 weeks post inoculation) in POLE1-silenced (TRV-NbPOLE1) or control (TRV) N. benthamiana plants measured by qPCR. Plants inoculated with the empty vector (EV) are used as negative control. Data are the mean of five independent biological replicates; error bars represent SD. The 25S ribosomal DNA interspacer (ITS) was used as a reference gene; values are presented relative to ITS. All experiments were repeated at least three times with similar results, with the exception of the ChIP assays, which were repeated twice. Two-sided Student’s t test (f, g, h, I, j, k) was performed to test statistical significance (****P < 0.0001; ***P < 0.001; **P < 0.01). ns: not significant. The original data from all experiments and replicates can be found in the Source data file.