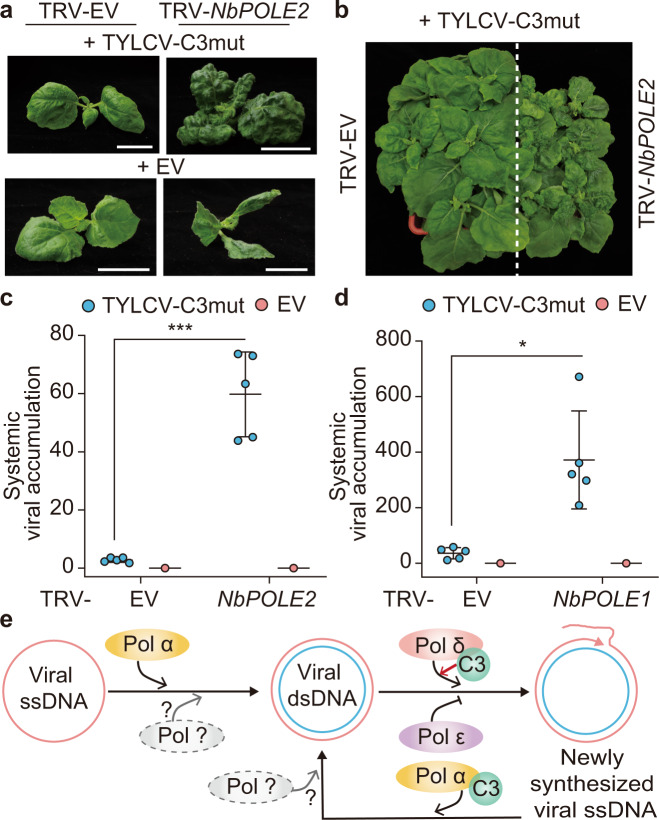
Fig. 4. Silencing subunits of DNA polymerase ε enables systemic infection of a C3 null mutant geminivirus.
a, b Symptoms of POLE2-silenced (TRV-NbPOLE2, right) or control (TRV-EV, left) N. benthamiana plants inoculated with a C3 null mutant TYLCV at 4 weeks post inoculation. EV: empty vector control. Scale bar: 2 cm. c, d Viral accumulation in systemic TYLCV infections (4 weeks post inoculation) in POLE2-silenced (TRV-NbPOLE2) (c), POLE1-silenced (TRV-NbPOLE1) (d), or control (TRV-EV) N. benthamiana plants measured by qPCR. Plants inoculated with the empty vector (EV) are used as negative control. Data are the mean of five independent biological replicates; error bars represent SD. The 25S ribosomal DNA interspacer (ITS) was used as a reference gene; values are presented relative to ITS. Asterisks indicate a statistically significant difference according to a two-sided Student’s t test (***P < 0.001; *P < 0.05). e Hypothetical model of the role of DNA polymerases α and δ in the replication of the geminiviral genome. DNA polymerase α is required to convert the viral ssDNA genome to the dsDNA replicative intermediate, which is then replicated by DNA polymerase δ to produce new viral ssDNA. The virus-encoded C3 protein interacts with DNA polymerase α (POLA2) and DNA polymerase δ (POLD2), and selectively recruits the latter over the non-productive DNA polymerase ε. The original data from all experiments and replicates can be found in the Source data file.