Figure 1.
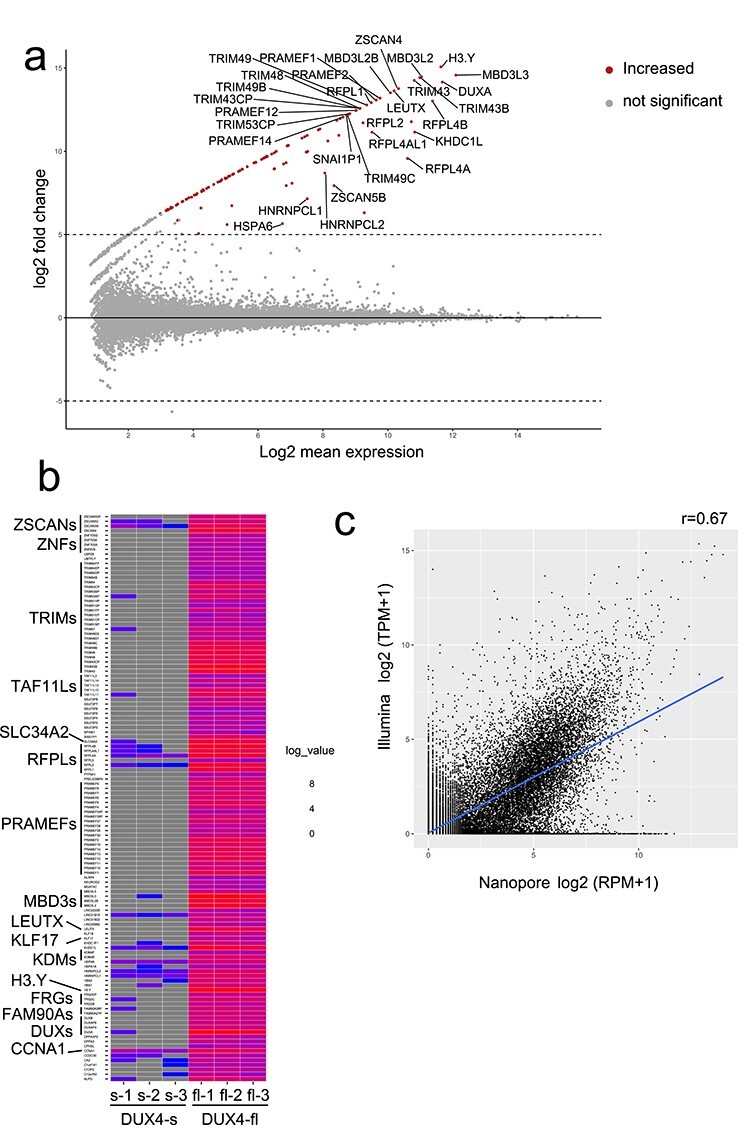
(a) MAplot for differentially expressed genes (DEGs) in triplicated nanopore dRNA-seq data from DUX4-fl and DUX4-s overexpressing RD cells. Only the top 30 genes are shown. The transcripts with adjusted P-value <0.001 and log2fold change ≥5 were determined as differentially expressed transcripts. (b) Heatmap of DEGs. Well-known DUX4 target genes and TAF11-like pseudogenes were significantly upregulated by DUX4-fl overexpression. (c) Correlation between Illumina short-read cDNA-seq and Nanopore long-read dRNA-seq expression in DUX4-fl-induced genes. Pearson correlation coefficients are shown.
