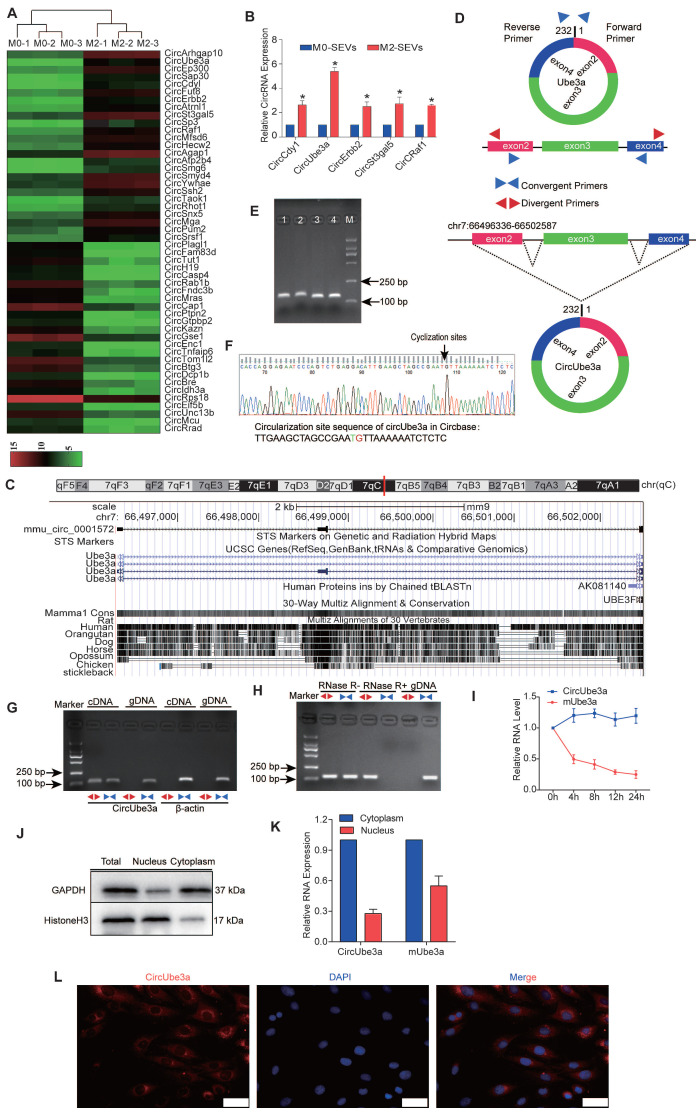
Figure 4.
Characteristics of circUbe3a carried in M2M-SEVs. A: Differentially expressed circRNAs in M0M-SEVs and M2M-SEVs, as identified by microarray analysis. B: Differentially expressed circRNAs in M0M-SEVs and M2M-SEVs were further analyzed by RT-qPCR (*P < 0.05 vs M0M-SEVs). C: CircUbe3a is located in the genome at chr7:66496336-66502587. A schematic diagram of circUbe3a formation is shown. D: Schematic diagram showing the positions of the divergent and convergent primers. E: Nucleic acid electrophoresis band. The molecular weight of the PCR product was approximately 100 bp. F: The sequence of circUbe3a in circBase (upper part) was consistent with the Sanger sequencing results (lower part). G: Nucleic acid electrophoresis revealed the PCR products amplified by the divergent and convergent primers. H: Nucleic acid electrophoresis revealed the PCR products before and after RNA enzyme treatment. I: The half-lives of circUbe3a and linear mUbe3a were measured after treatment with actinomycin D. J: RNAs and their encoded proteins in the cytoplasm and nucleus were extracted and isolated using a nucleocytoplasmic separation kit, and then Western blot analysis was performed to assess protein expression to evaluate the separation efficiency. K: qPCR was used to detect the nucleoplasmic expression of Ube3a linear mRNA and circRNA. L: RNA-FISH with Cy3-labeled probes was conducted to detect circUbe3a expression in CFs. Nuclei were stained with DAPI. Scale bar = 20 μm.