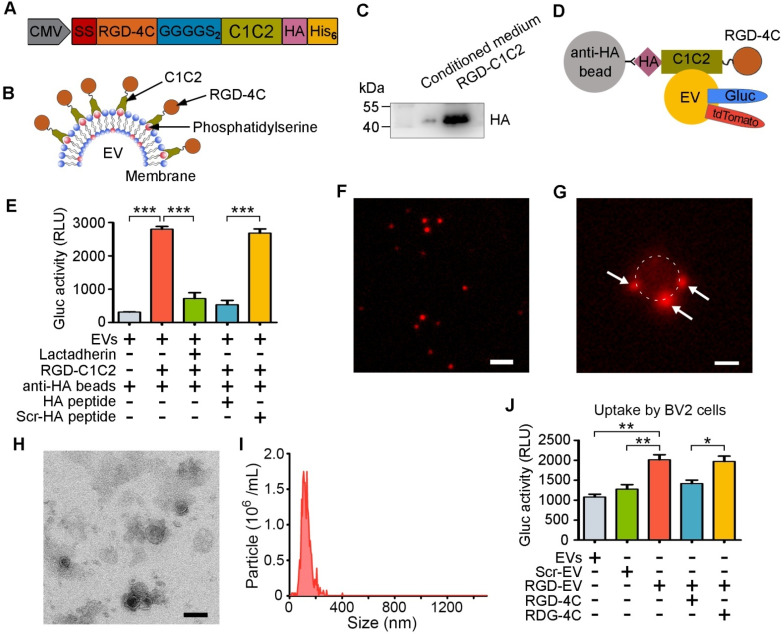
Figure 2.
Preparation and characterization of RGD-EVReN. (A, B) Schematic diagram of designing RGD-C1C2 and decorating EVs with RGD-C1C2. (A) Recombiant protein consists of a signal sequence (SS) for secreting, RGD-4C peptide sequence, 2×GGGGS linker sequence, C1C2 domains of lactadherin, and HA and His6 tags. (B) C1C2 domains of the recombiant protein bind phosphatidylserine on EV surface, resulting in decoration of RGD-4C. (C) Western blots of purified RGD-C1C2 and corresponding conditioned medium, stained by anti-HA antibody. (D) Schematic diagram of EV pull-down assay. tdTomato or Gluc-labeled RGD-EVReN are expected to be pulled-down by anti-HA beads through affinity to HA-tag on RGD-C1C2. (E) Anti-HA beads were applied to pull-down Gluc-displayed EVReN with RGD-C1C2-association or not followed by luminescence detection (n = 4). For blocking assays, EVReN were preincubated with lactadherin, or the beads were preincubated with the HA peptide or the scrambled HA peptide (Scr-HA). (F) tdTomato-labeled RGD-EVReN present small particles (Red) on fluorescent image. Scale bar, 2 µm. (G) RGD-EVReN (Red particles) appear on the beads after being pulled-down. The bead is around by dashed line. Scale bar, 1 µm. (H) TEM image of RGD-EVReN. Scale bar, 100 nm. (I) Size distributions of RGD-EVReN based on NTA measurements. (J) Luminescence detection of cellular uptake (n = 4) of Gluc-displayed EVReN or Scr-EVReN or RGD-EVReN after 2-h incubation with BV2 microglia. For blocking assays, cells were preincubated with the RGD-4C peptide or the scrambled peptide (RDG-4C). All the quantitative data are expressed as mean ± SEM. **P < 0.01, ***P < 0.001 by One-way ANOVA.