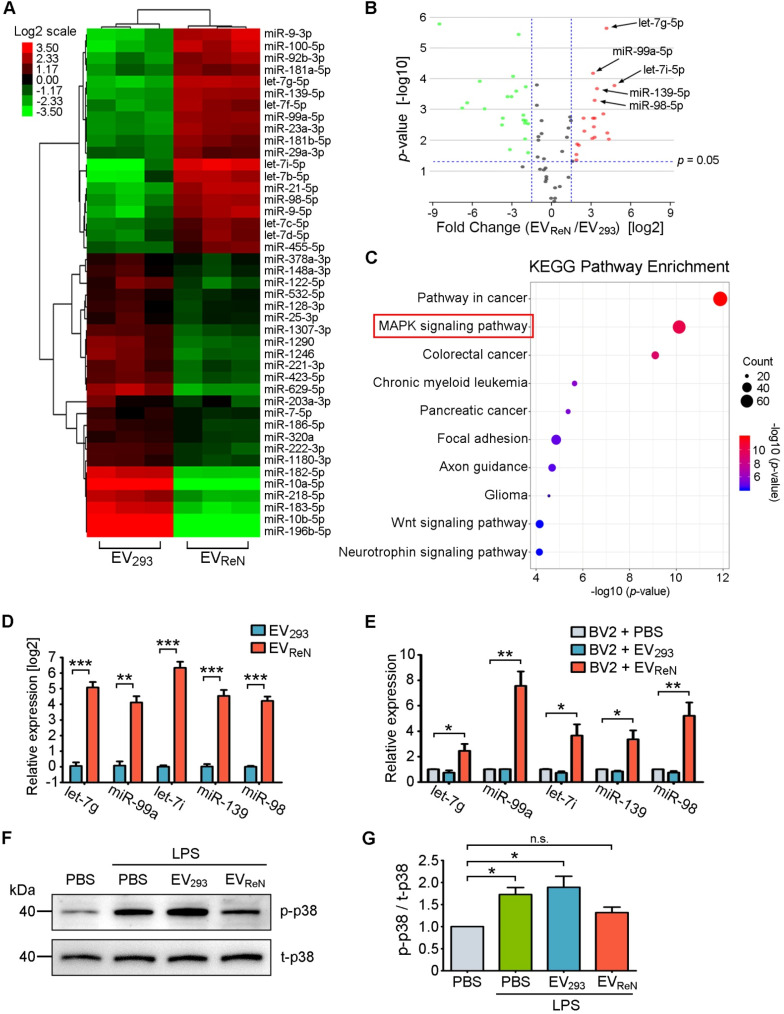
Figure 6.
miRNAs enriched in EVReN inhibit MAPK pathway. (A) Clustering and Heatmap analysis (n = 3) of 43 differentially packaged miRNAs in EV293 and EVReN (TPMaverage > 1000, fold change > 1.5 and p < 0.05). (B) Volcano plot shows the relation between the p-values and the fold changes. (C) KEGG pathway analysis of the target genes of 7 significantly up-regulated miRNAs in EVReN. Top 10 enriched pathways are indicated. (D) RT-PCR analyses (n = 3) of top 5 differently packaged miRNAs in EV293 and EVReN. cel-miR-39 as the external reference. (E) RT-PCR analyses (n = 3) of the miRNAs in BV2 cells were performed 24 h after the treatment of PBS, EV293 or EVReN. U6 as the internal reference. (F) BV2 cells were pretreated with PBS, EV293 or EVReN for 24 h before LPS treatment. The expression levels of phosphorylated p38 (p-p38) and total p38 (t-p38) in the cells were determined by Western blot analysis 24 h after LPS treatment. (G) Quantification of p-p38 (n = 3) was normalized to t-p38 and presented as a relative change compared with the group without LPS treatment. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., no significance versus the first groups by One-way ANOVA.