Figure 1.
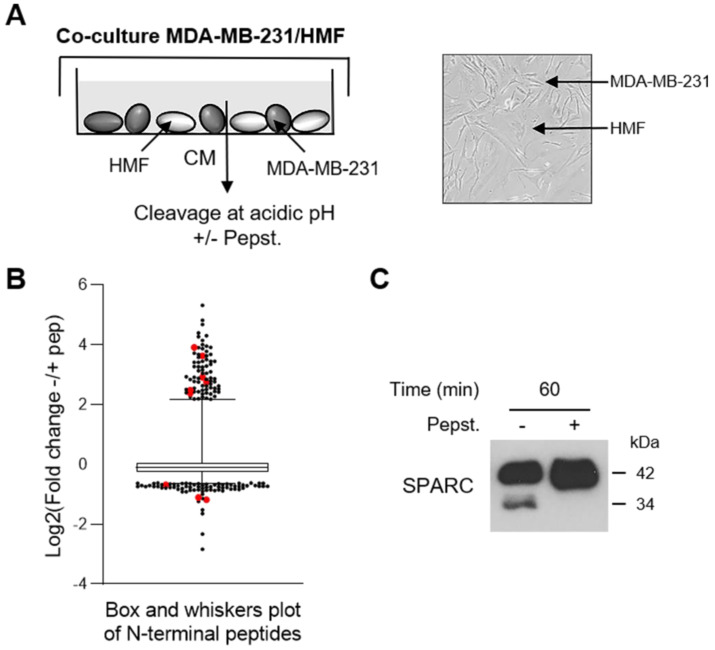
Identification of SPARC as an extracellular protein cleaved in the TNBC microenvironment at acidic pH. (A) Experimental set-up of the MDA-MB-231 cell-HMF co-culture system. MDA-MB-231 TNBC cells and HMFs were co-cultured in serum-free DMEM without sodium bicarbonate and phenol red and buffered with 50 mM HEPES [pH 7.5] at 37 °C for 24 h. The 24 h-conditioned medium was then concentrated to 0.2 mg/mL, and incubated in cleavage buffer with or without pepstatin A (Pepst.) (12.5 µM) at pH 5.5 and 37 °C for 60 min. A representative image of an MDA-MB-231/HMF co-culture is shown in the right panel (x 200). (B) Box and whisker plot of the normalized ratios of the N-terminal peptides identified by TAILS in the MDA-MB-231/HMF co-culture secretome. 3091 peptides quantified at t = 60 min of incubation with cleavage buffer were used to generate the graph. The without/with pepstatin A ratios corresponding to SPARC peptides are highlighted in red; whiskers correspond to the 2.5th and 97.5th percentiles. (C) Validation of SPARC cleavage in the MDA-MB-231/HMF co-culture secretome. Secretome samples (2 µg) from the MDA-MB-231/HMF co-culture incubated in cleavage buffer with or without pepstatin A (Pepst.) (12.5 µM) at pH 5.5 and at 37 °C for 60 min were separated on 13.5% SDS-PAGE followed by immunoblotting with anti-SPARC antibody (clone AON-5031).