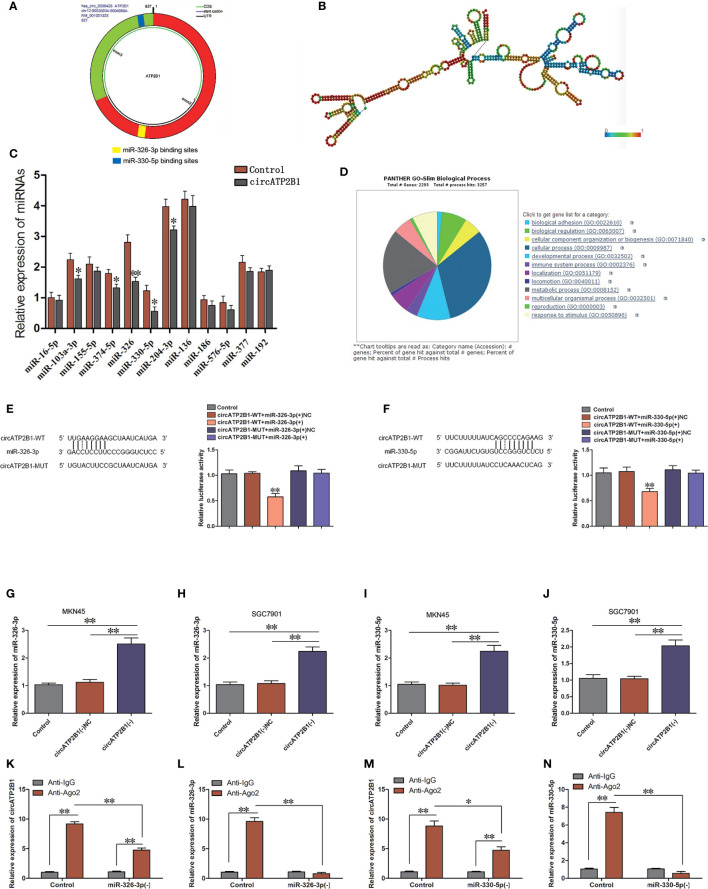
Figure 2.
circATP2B1 served as a sponge for miR-326-3p and miR-330-5p in gastric cancer cells. (A). Shematic illustration showing miR-326-3p and miR-330-5p binding sites of circATP2B1. (B). Secondary structure of circATP2B1. (C–F) The predicted matching sequence of miR-326-3p (C) and miR-330-5p (E) to circATP2B1. (circATP2B1-Wt) and the designed mutant sequence (circATP2B1-Mut) were indicated. (D, F) Luciferase assay of circATP2B1 and miR-326-3p and miR-330-5p was conducted. Relative luciferase activities for circATP2B1-Wt and circATP2B1-Mut group were presented. Data were presented as mean ± SD. (*P <0.05, **P <0.01). (G–J). Relative expression of miR-326-3p (G, H) and miR-330-5p (I, J) was investigated in MKN45 and SGC7901 cells using qRT-PCR after circATP2B1 silencing. (K–N). miR-326-3p/miR-330-5p was identified in the circATP2B1-RISC complex. Relative expression of circATP2B1 (K), miR-326-3p (L), circATP2B1 (M), miR-330-5p (N) were measured with qRT-PCR. Data represent means ± S D (n = 3 for each group), (*P <0.05, **P <0.01).