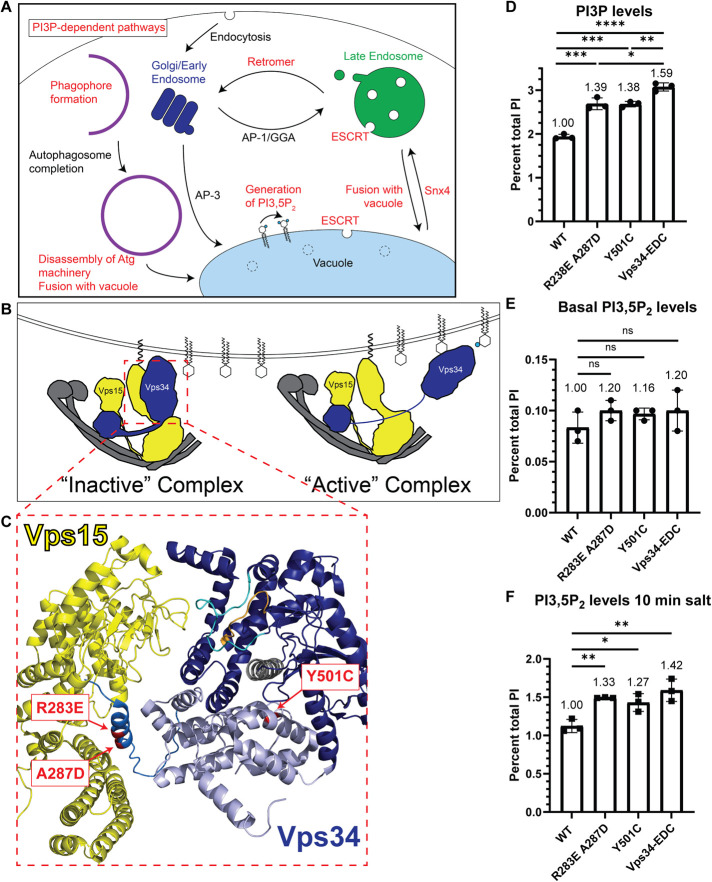
FIGURE 1:
Generation of hyperactive Vps34 mutants. (A) Schematic indicating several PI3P-dependent intracellular trafficking pathways in yeast. PI3P serves as the substrate for PI(3,5)P2. PI3P is also required for Snx4-dependent retrograde transport from the vacuole and retromer-dependent retrograde transport from endosomes. ESCRT function at late endosomes and the vacuole is also dependent on PI3P. Furthermore, PI3P is required for phagophore formation during the initiation of autophagy, and then PI3P is removed before the disassembly of some autophagy proteins from the surface of mature autophagosomes. In addition, fusion of autophagosomes and other vesicles with the vacuole, as well as homotypic vacuole fusion (not depicted), are PI3P-dependent pathways. (B) Vps34 is proposed to be regulated in part via changes in contact of the Vps34 HELCAT domain with Vps15. In the inactive conformation, the Vps34 HELCAT domain contacts the Vps15 scaffold. During activation, the Vps34 HELCAT domain is proposed to alter the contact with Vps15 and allow Vps34 to access its PI substrate (Stjepanovic et al., 2017). (C) Crystal structure of the helical (light blue) and kinase (dark blue) domains of Vps34 and its contact with the pseudokinase domain of Vps15 (yellow; Rostislavleva et al., 2015). The three amino acid changes that comprise the Vps34-EDC hyperactive mutant are indicated (red). Two of these mutations, R283E and A287D, are on an alpha-helix N-terminal to the helical domain of Vps34 (neutral blue) and may hinder Vps34 HELCAT interaction with Vps15 and favor the active Vps34 conformation. The Y501C mutation is in the helical domain of Vps34 and faces the alpha-C helix (gray) of the kinase domain, nearby the activation (cyan) and catalytic (orange) loops of Vps34. (D–F) The Vps34 mutants R283E A287D and Y501C elevate PI3P levels. PI(3,5)P2 levels are also elevated during hyperosmotic shock. Combining the mutants to Vps34-EDC further elevated PI3P levels and PI(3,5)P2 levels. vps34Δ cells were transformed with a wild-type or mutant pRS416-Vps34 plasmid. PPI lipid levels were measured by metabolically labeling cells with myo-3H-inositol for 16 h. Prior to harvest, indicated cultures were exposed to 10 min of hyperosmotic shock. PPI lipid head groups were separated by anion exchange and HPLC. n = 3. Error bars indicate SD. Unpaired t test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.