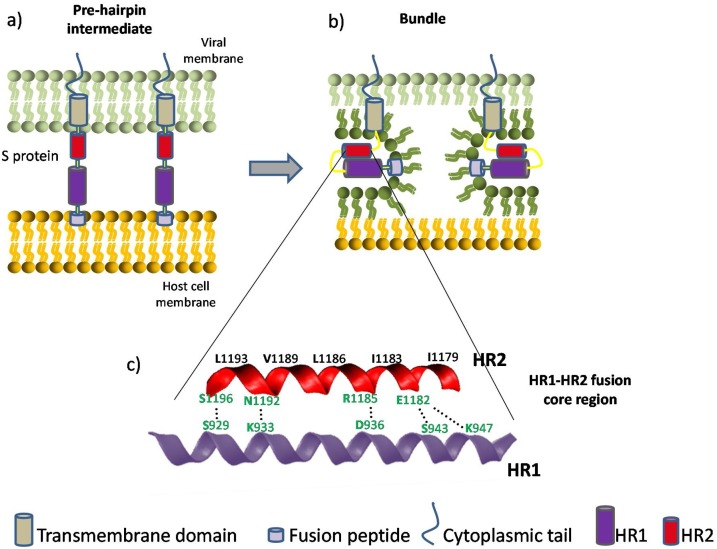
Fig. 1.
Coronavirus viral fusion pathway based on class I fusion protein model. For simplicity, only two stages are depicted. After the S protein achieves a pre-fusion metastable state, relevant proteases enable the fusion peptide to insert in the host membrane and allow the S protein to form the pre-hairpin intermediate (a). The pre-hairpin begins to fold back on itself due to HR1 and HR2 interactions forming the bundle (b). During the S protein foldback, the two membranes will approach each other until the outer leaflets merge and eventually the inner leaflets merge (pore formation). Adapted from [99,100]. (c) Cartoon representation and closer look of the HR1-HR2 fusion core region of SARS-CoV-2. The HR motifs consist of a group of tandemly arranged seven-residue repeats. This sequence feature has the capacity to generate a hydrophobic interface along an α-helix, enabling the formation of a coiled-coil structural module when several HR-containing polypeptide chains are arranged together in parallel [101]. This fusion core is a particularly well studied target for antiviral peptides, as they can be designed in order to competitively inhibit this interaction. Relevant binding residues are labeled. Among the important amino acids responsible for the HR1-HR2 interaction, the residues colored in green may contribute to the enhanced interactions between HR1 and HR2 and stabilize 6-HB conformation of SARS-CoV- 2 compared with those of SARS-CoV (adapted from [34]).