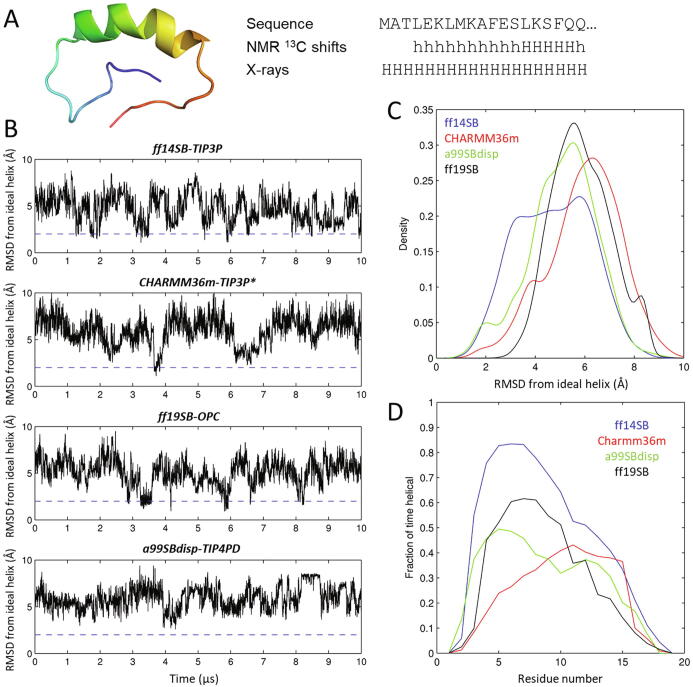
Fig. 2.
Tests on Huntingtin’s Nt19 peptide. (A) Model of Huntingtin segment Nt17Q17 computed from 13C chemical shifts at neutral pH by Baias et al. [46], showing on the right the sequence of the Htt-1-19 peptide under study and the helical propensities derived for it from 13C chemical shifts [45] and from an X-ray structure of huntingtin’s exon 1 fused to maltose binding protein (PDB ID 3IO6), where “h” and “H” mean low and helical propensity, respectively. (B) RMSD from an ideal helical conformation over 10 μs of unbiased MD simulation of Htt-1-19. The dashed line is set at 2 Å RMSD, under which the peptide is essentially fully helical. (C) Histogram of RMSD densities from the data in panel B. (D) Fractional helicity per residue averaged throughout each of the four simulations. The total average helical content is 45% for ff14SB-TIP3P, 23% for CHARMM36m-TIP3P*, 28% for a99SBdisp-TIP4PD and 29% for ff19SB-OPC.