Figure 9: MolDarting does not improve sampling when the simulation moves outside the darts.
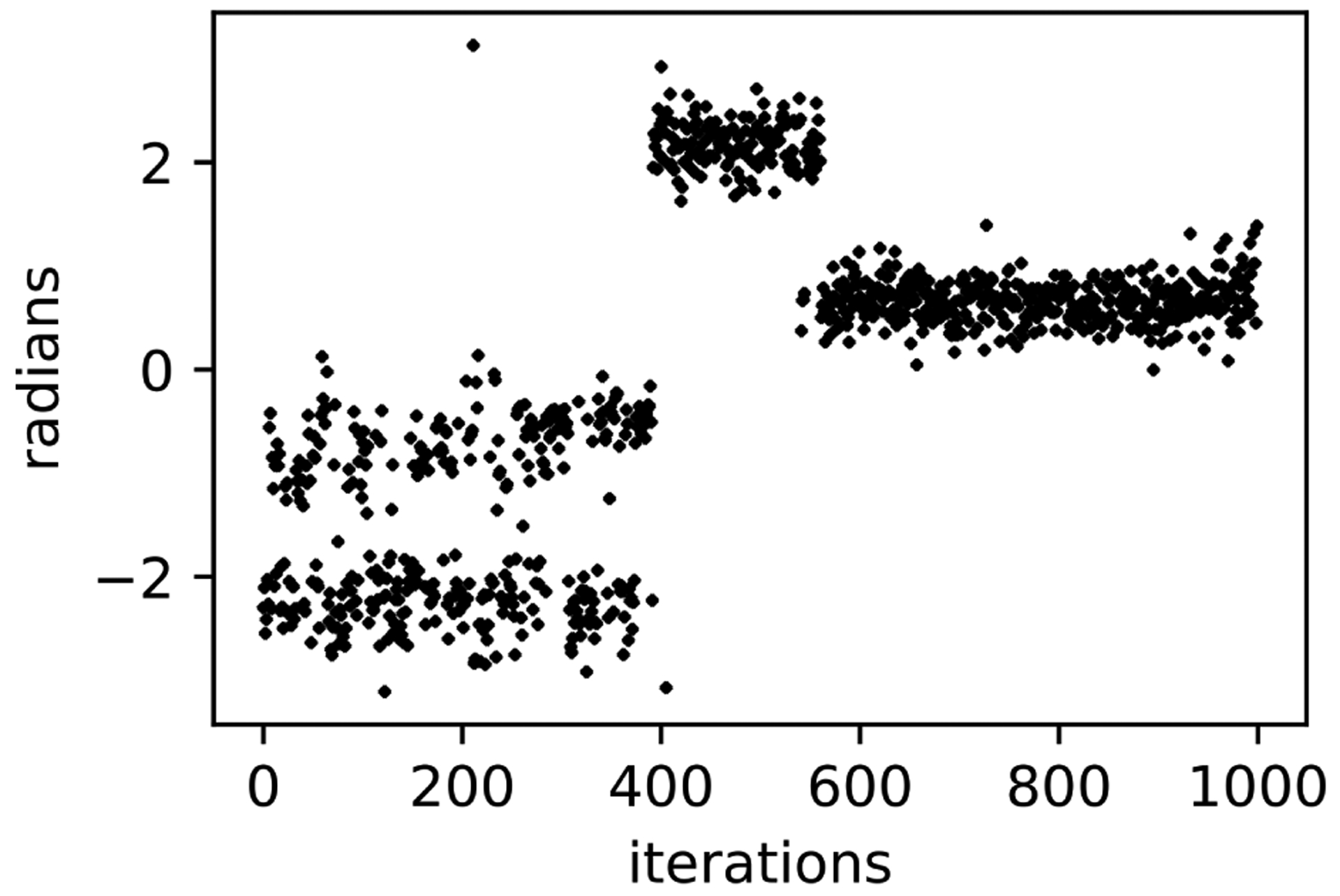
Here, the initial binding modes of toluene between 0 and π radians are well sampled (in the first 400 iterations), since these are covered by the rotational regions from MolDarting. However if the simulation leaves that region, then a MolDarting move cannot take place, and thus the simulation becomes just a normal MD simulation. In this particular simulation, around the 400th iteration toluene flips to the symmetric equivalent binding mode, which is not covered by the rotational regions, greatly reducing sampling.
