Figure 3.
Results from global fit of all sample-peptide pairs with applied mlxp cutoff
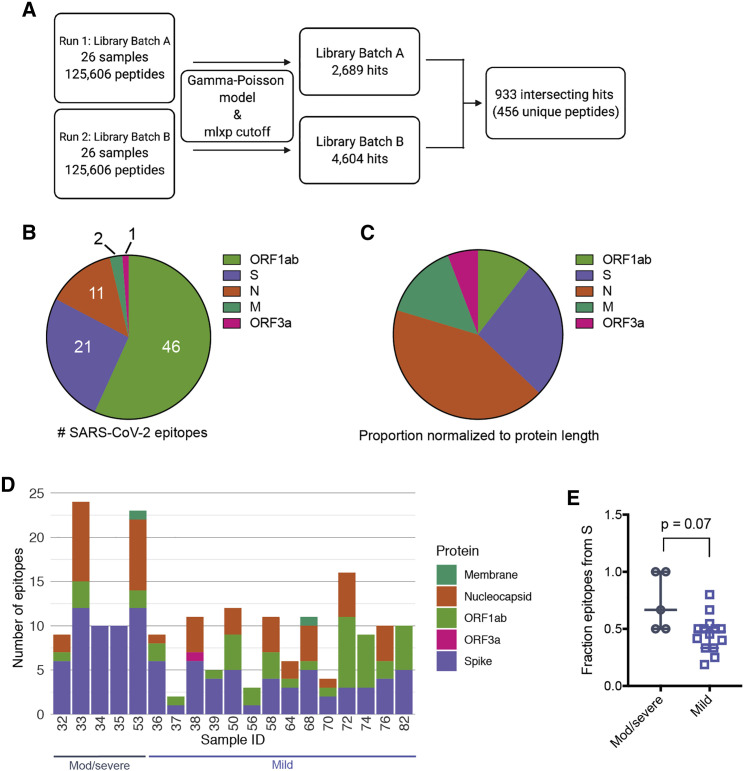
(A) Data-processing scheme. Samples were tested with two separate phage libraries (library 1 and library 2; Figure S1). Peptide enrichment was scored using a gamma-Poisson model, and data were curated using a cutoff corresponding to FPR 0.05 (Figure S1).
(B) Proportion of SARS-CoV-2 epitopes derived from individual proteins in all patient samples tested. Numbers indicate the total enriched SARS-CoV-2 epitopes from each ORF.
(C) Proportions in (B) normalized with respect to polypeptide length.
(D) Epitope counts across COVID-19 patient samples for SARS-CoV-2 only. Bars are further sectioned by SARS-CoV-2 ORF, indicated to the right.
(E) Fraction of total epitopes arising from the S protein, calculated for moderate/severe and mild samples (number of S epitopes/number of total epitopes). p value was calculated using a two-tailed unpaired Welch’s t test (n = 5, moderate/severe COVID-19, n = 14 mild COVID-19; bars represent median and interquartile range).