Figure 5.
SARS-CoV-2 cross-reactivity in two populations, SARS-CoV-2 unexposed and COVID-19
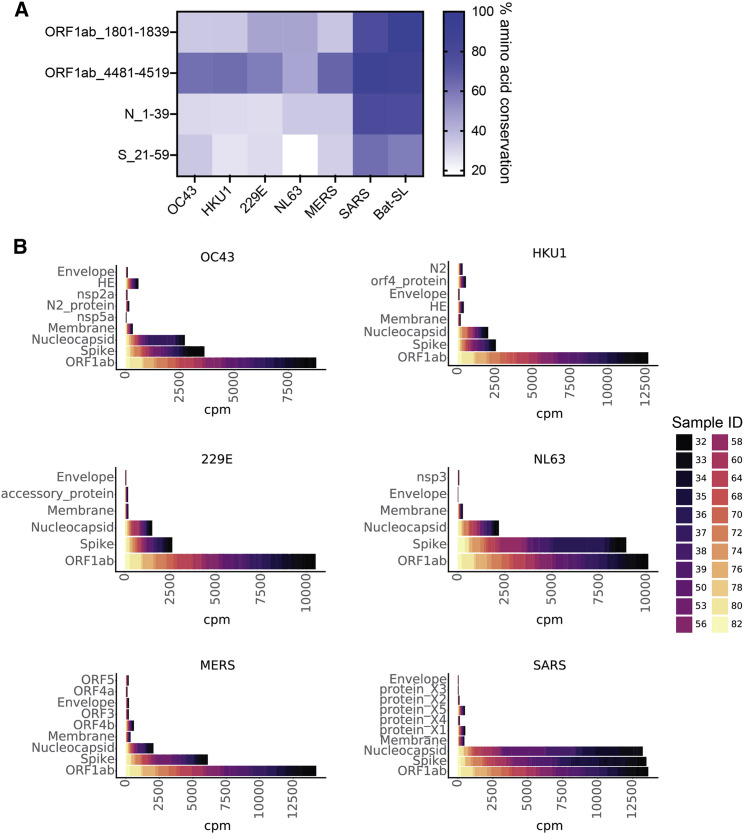
(A) Four SARS-CoV-2 epitopes that were reactive in unexposed/pre-pandemic individuals are shown on the vertical axis. The heatmap displays the percentage of amino acid conservation after local alignment with representative strains of the other circulating CoVs in the pan-CoV phage library.
(B) Stacked bar plots showing CPM (cpm) for peptides from each of the viral proteins on the y axis for each of the six non-SARS-CoV-2 HCoVs in the phage library. Colors represent individual samples, as indicated by the legend on the right. Representative endemic HCoV strains used in (A) and (B) are OC43_SC0776, HKU1_Caen1, 229E_SC0865, and NL63_ChinaGD01. Protein names on y axes are identical to GenBank entries for each viral protein (see STAR Methods).