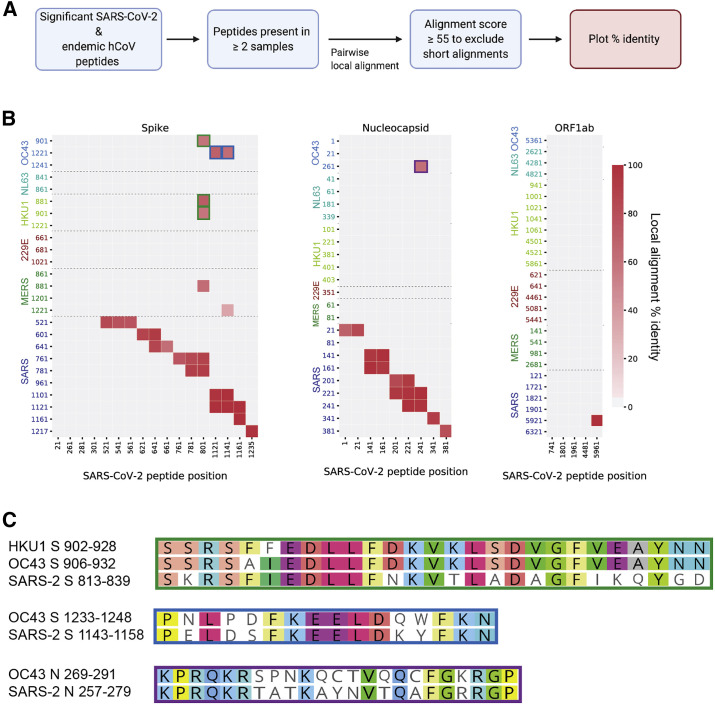
Figure 6.
Homology among significant HCoV/SARS-CoV-2 sequence pairs in individuals with COVID-19
(A) Unique peptide hits from all CoVs that were present in two or more COVID-19 patient samples were subjected to Smith-Waterman local alignment. Sequences that were 100% identical between SARS-CoV-2 and the other CoVs were not included in the analysis.
(B) Peptide pairs with alignment scores >55 (Figure S5) were plotted to show percent identity. Peptide start positions from SARS-CoV-2 are listed on the x axis, and peptide start positions from the other human-infecting CoVs are listed on the y axis. Green, blue, and purple outlines match with the corresponding peptides pairs shown in (C).
(C) Local sequence alignments for the high-scoring peptide pairs in (B).