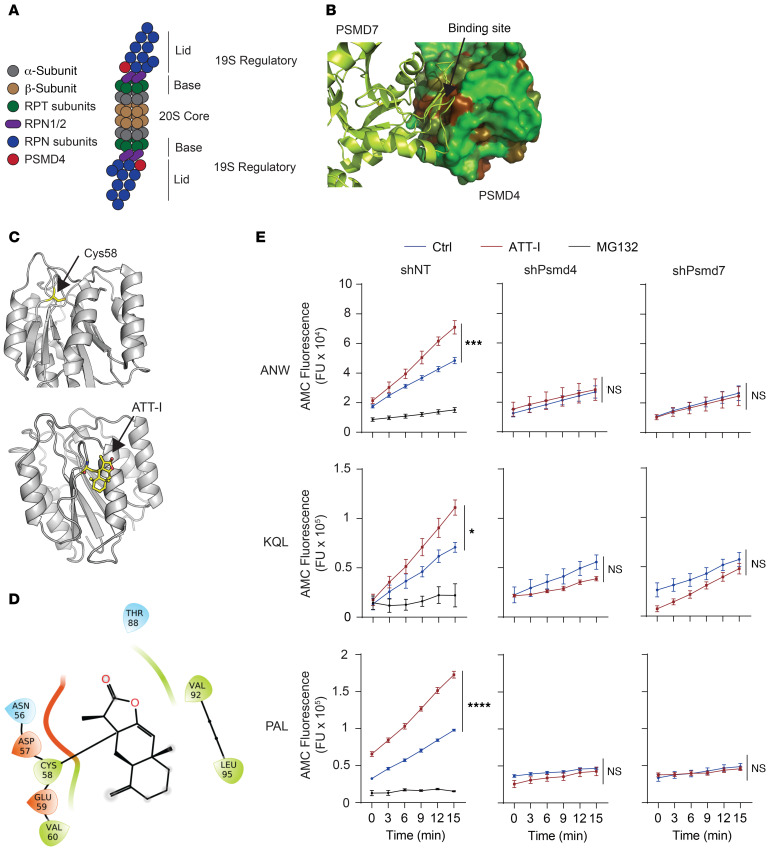
Figure 4. ATT-I binds to PSMD4 and stabilizes the PSMD4 and PSMD7 interaction, leading to enhanced proteasomal activities.
(A) Schematic representation of the immunoproteasome. (B) Three-dimensional structure of the complex between PSMD4 and PSMD7 obtained from the cryo-EM structure of the 26S proteasome (PDB code 6EPD). PSMD4 is shown in solvent-accessible surface area, color-coded based on hydrophobicity (brown is hydrophobic and green hydrophilic). PSMD7 is shown in green ribbon representation. (C) Three-dimensional structure of PSMD4 shown in gray ribbon representation. The cysteine residue Cys58 located at the PSMD4 and PSMD7 interface is shown in capped-sticks rendering (upper panel). The predicted structure of the covalent complex between PSMD4 and ATT-I (bottom panel). PSMD4 is shown in gray ribbon rendering, and Cys-58 and ATT-I are depicted in capped-sticks representation (yellow, red, blue, and gold correspond to carbon, oxygen, nitrogen, and sulfur, respectively). (D) Ligand interaction diagram showing individual interaction of ATT with neighboring amino acids on PSMD4. (E) Activity analysis of immunoproteasomes purified from control or PSMD4-knockdown MC38 cell lysates upon treatment with ATT-I using different substrates (ANW, KQL, and PAL) as indicated. Quantitative data are presented as mean ± SD of 2 parallel experiments (n = 2). Statistical analysis was conducted using 2-way ANOVA. *P < 0.05; ***P < 0.001; ****P < 0.0001.