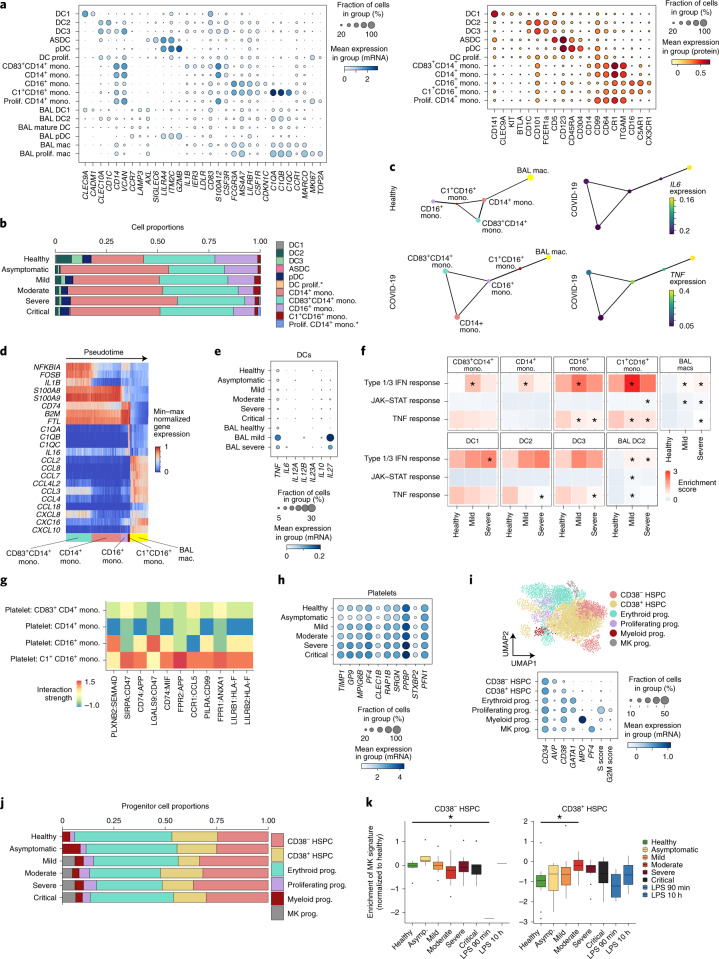
Fig. 2. Expansion of complement-expressing nonclassical monocytes and megakaryocyte-primed progenitor cells and increased platelet activation with COVID-19 disease severity.
a, Dot plots of gene (left) and surface protein (right) expression for myeloid populations. b, Bar plot of the proportion of myeloid populations from the Newcastle and London sites. Hypothesis testing was performed using a quasi-likelihood F-test comparing healthy controls to individuals with COVID-19. Differentially abundant cell types were determined using a 10% FDR and are marked with an asterisk. c, PAGA graph representing connectivity between clusters defined in a for healthy (top left) and COVID-19 (bottom left) monocytes and BAL macrophages (mac). Expression of IL6 (top right) and TNF (bottom right) in each cluster along the predicted path for COVID-19 monocytes. d, Expression of differentially expressed cytokines between CD83+CD14+ monocytes and BAL macrophages shown by cells ordered by pseudotime calculated for cells from c. e, Dot plot of gene expression of DC-derived T cell polarizing cytokines in peripheral blood DC2 cells and mature BAL DCs. f, Heat map displaying gene-set enrichment scores for type 1/3 IFN response, TNF response and JAK–STAT signatures in the myeloid populations. Asterisks indicate significance compared to healthy controls. Absolute values and other comparisons are provided in Supplementary Table 7. g, Heat map of predicted ligand–receptor interactions between platelets and monocyte subsets, using RNA data. h, Dot plot of significant differentially expressed genes between samples from healthy donors and individuals with COVID-19 filtered for platelet activation markers. i, UMAP representation of HSPCs (top) and dot plot of gene expression markers used to annotate clusters (bottom). MK, megakaryocyte; prog, progenitor. j, Bar chart depicting the proportion of progenitors. k, Box plots displaying the enrichment of a megakaryocyte progenitor signature in CD34+CD38+ HSPCs (right) and CD34+CD38− (left), averaged per donor scores. Comparisons were made by an analysis of variance (ANOVA) with pairwise comparisons using Tukey’s test. Asterisks above bars indicate significance and are colored by the severity for which they were compared to. Absolute values are provided in Supplementary Table 8. Boxes denote the interquartile range (IQR), and the median is shown as horizontal bars. Whiskers extend to 1.5 times the IQR, and outliers are shown as individual points (P values: CD38-negative cells in healthy versus LPS group (90 min), 0.3 × 10−3; CD38-positive cells in healthy versus moderate group, 0.7 × 10−3).