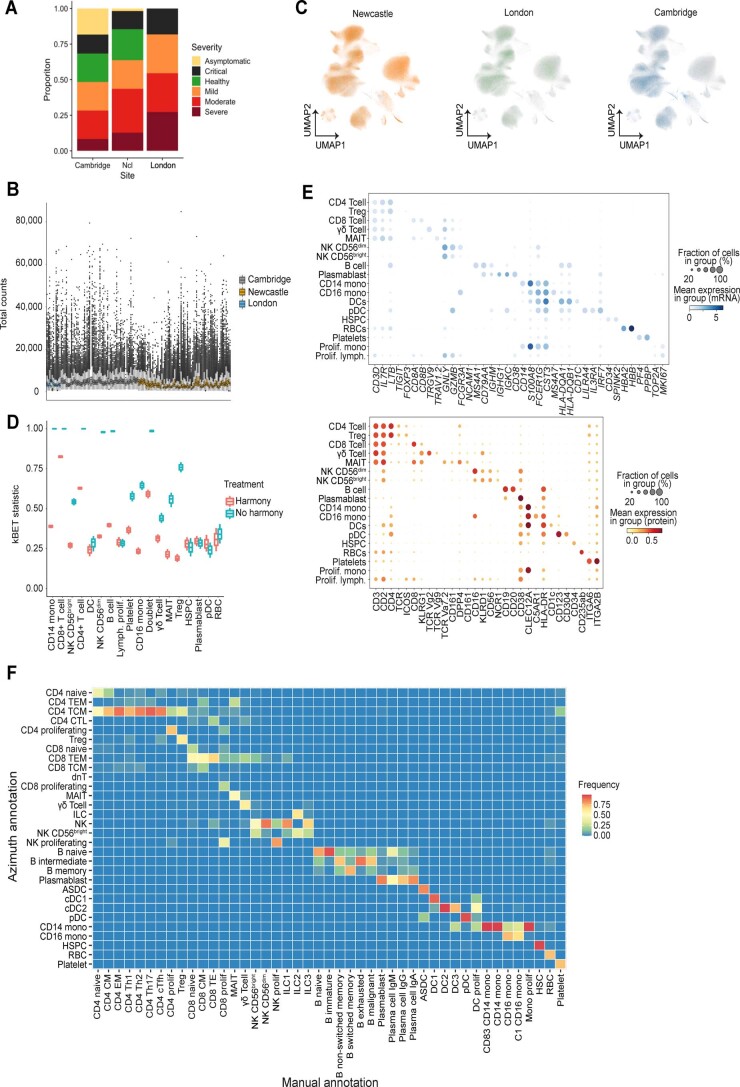
Extended Data Fig. 1. Single-cell analysis quality control and cell type definition for COVID PBMC single cell analysis.
a, Bar chart showing the composition of sample severities across the three sites (n = 23 mild, n = 30 moderate, n = 11, severe, n = 10 critical biologically independent samples). b, Scatter plot displaying the total number of gene counts per sample from each site. c, UMAP visualisations from Fig. 1b coloured by site. d, Boxplot of kBET results calculated both before and after batch correction with Harmony for each cluster in Fig. 1b kBET statistic calculation using patient ID as the batch factor (n = 130 biologically independent samples, n = 627,172 cells in 1 experiment). Hinges indicate to 25th and 75th percentile and whiskers to lowest and highest value in 1.5*interquartile range. e, Dot plots of 5’ gene expression (top; blue) and surface protein (bottom; red) expression for populations shown in Fig. 1a where the colour is scaled by mean expression and the dot size is proportional to the percent of the population expressing the gene/protein, respectively. f, Tile plot showing percentage concordance between COVID-19 PBMC annotation (x-axis) and Azimuth annotation (y-axis) (https://satijalab.org/azimuth/).