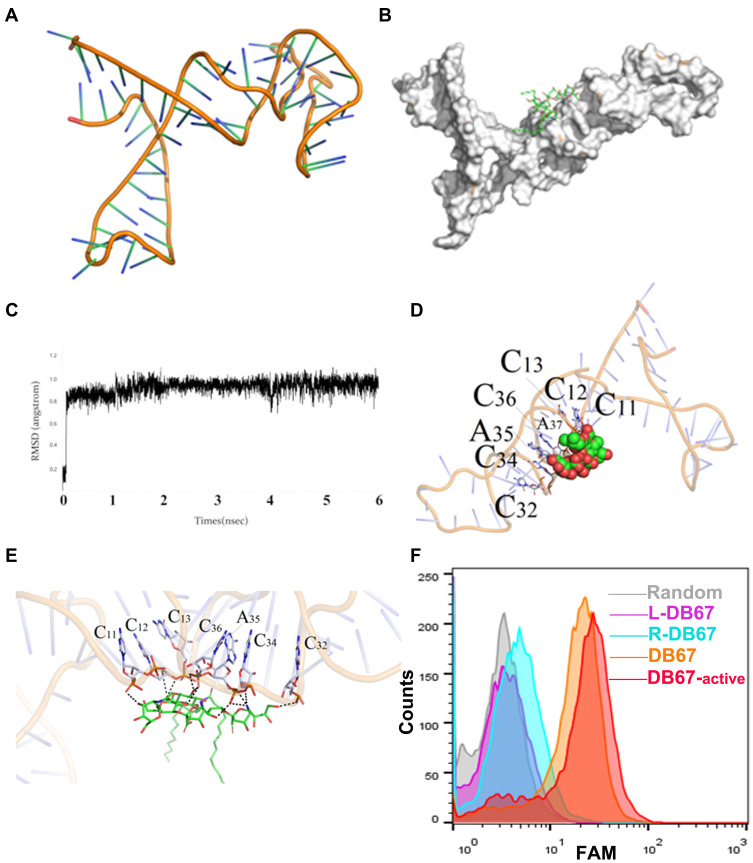
Figure 3.
Evaluation of DB67 core region for GD2 recognition and binding. (A) The three-dimensional of DB67. DB67 was mainly composed of several stem-ring structures. (B) Black box docking position of DB67. (C) Recognition of system balance between DB67 and GD2 molecules. (D and E) Molecular docking. The C11th, C12th, C13th, C32th, C34th, A35th, and C36th could form polar interaction. (F) Flow cytometry assessment of truncated DB67 for GD2 binding specificities. DB67 was truncated and truncated sequence L-DB67, R-DB67, DB67-active, and DB67 were incubated with IMR32 cells. Fluorescence signals were detected.