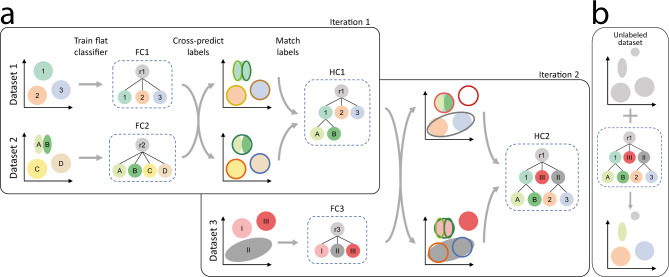
Fig. 1. Schematic overview of scHPL.
a Overview of the training phase. In the first iteration, we start with two labeled datasets. The colored areas represent the different cell populations. For both datasets a flat classifier (FC1 and FC2) is constructed. Using this tree and the corresponding dataset, a classifier is trained for each node in the tree except for the root. We use the trained classification tree of one dataset to predict the labels of the other. The decision boundaries of the classifiers are indicated with the contour lines. We compare the predicted labels to the cluster labels to find matches between the labels of the two datasets. The tree belonging to the first dataset is updated according to these matches, which results in a hierarchical classifier (HC1). In dataset 2, for example, subpopulations of population “1” of dataset 1 are found. Therefore, these cell populations, “A” and “B”, are added as children to the “1” population. In iteration 2, a new labeled dataset is added. Again a flat classifier (FC3) is trained for this dataset and HC1 is trained on datasets 1 and 2, combined. After cross-prediction and matching the labels, we update the tree which is then trained on all datasets 1–3 (HC2). b The final classifier can be used to annotate a new unlabeled dataset. If this dataset contains unknown cell populations, these will be rejected.