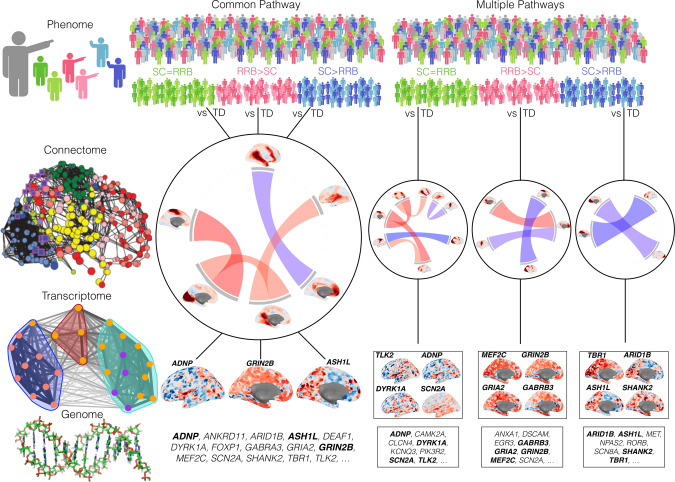
Fig. 1. Approach towards testing common pathway versus multiple pathways explanations behind SC-RRB balance in autism.
In this figure we depict two alternatives for how SC-RRB balance subtypes (phenome level; SC = RRB, green; RRB > SC, pink; SC > RRB, blue) could be explained at the level of macroscale functional connectome phenotypes measured with rsfMRI (connectome level) and autism-associated functional genomic mechanisms (e.g., transcriptome and genome levels). Columns in this figure depict the common pathway (middle) and multiple pathways (right) models. The common pathway model predicts that when each subtype is compared to a typically-developing (TD) comparison group, they converge and share a common difference from TD in affected macroscale rsfMRI functional connectome phenotype. Underpinning this shared connectome phenotype are a myriad of differing functional genomic mechanisms. At the level of the transcriptome, we identify genes linked to macroscale functional networks by identifying genes whose spatial expression pattern across the brain is similar to the spatial topology of the macroscale functional network. This procedure generates a list of genes relevant for such macroscale networks and these lists are then tested for enrichment in autism-association functional genomic mechanisms. The gene list at the genome level represents an example of possible autism-associated genes that may (bold) or may not (non-bold) be linked to macroscale functional networks. In contrast to the common pathway model, the multiple pathways model would highlight that differential connectome phenotypes when compared to TD are unique to each subtype, and that each of these subtype-specific connectome phenotypes is underpinned by a differing set of autism-associated functional genomic mechanisms.