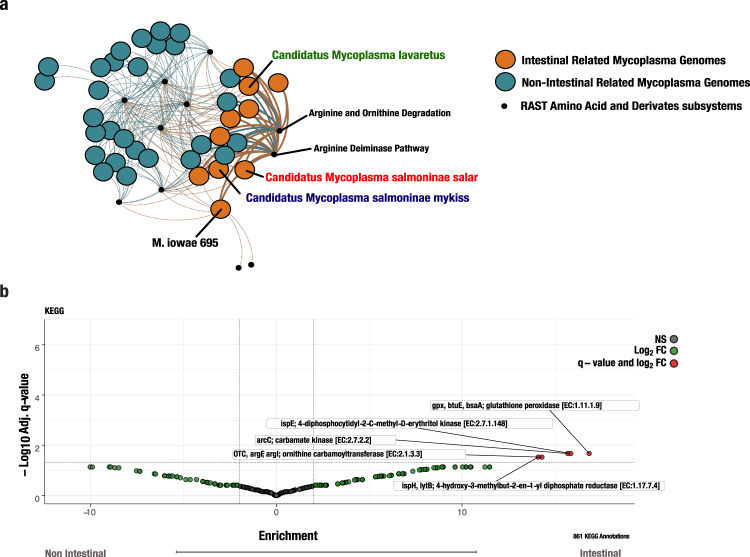
Fig. 4. Functional enrichment analysis between intestinal related Mycoplasma species and non-intestinal related Mycoplasma species.
a Genome clustering network based on the RAST ‘Amino Acid and derivatives’ subsystem. Clustering of genomes was based on RAST ‘Amino Acid and derivatives’ subsystems present among Mycoplasma genomes. The network layout was based on Force Atlas 2 algorithms. Blue colour indicates Mycoplasma genomes not being related to intestinal environments, whereas orange nodes indicate Mycoplasma genomes being related to intestinal environments. Small black nodes indicate RAST subsystems. Dashed edges indicate non-significant relations. Thick solid edges indicated significance between the RAST subsystem and intestinal environment. Blue, red and green font indicate the salmonid MAGs of Candidatus Mycoplasma salmoninae mykiss, Candidatus Mycoplasma salmoninae salar, and Candidatus Mycoplasma lavaretus, respectively. b Volcano plot of 861 KEGG annotated genes in Mycoplasma. Investigated genomes were divided into intestinal and non-intestinal related genomes of Mycoplasma according to Supplementary Data 2. Functional enrichment was inferred using a generalised linear model (GLMs) with the logit linkage function to compute an enrichment score and p value for each function. False detection rate correction to p-values to account for multiple tests was done and only genes with adjusted q-values below 0.05 and an enrichment score above 1 are reported.