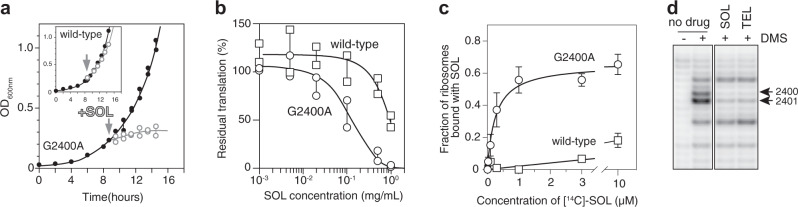
Fig. 2. Macrolide antibiotics bind to the mutant yeast ribosome and inhibit protein synthesis and cell growth.
a SOL arrests growth of the G2400A mutant S. cerevisiae cells. Plots show the growth of mutant cells in the absence of the drug (filled black circles) or after addition of 8× MIC of SOL (open gray circles). The inlet shows that wt yeast is resistant to SOL. The graphs represent results of two independent experiments with individual data points indicated. b Residual protein synthesis in wt (squares) and G2400A mutant (circles) yeast cells exposed to SOL. Translation was assessed by measuring incorporation of l-[35S]-methionine into polypeptides after exposure of cells for 10 min to different concentrations of SOL. l-[35S]-methionine incorporation in the untreated mutant cells was set to 100%. The data points of two independent experiments are indicated by squares (wt) and circles (G2400A mutant). c Equilibrium binding of [14C]-SOL to wt (squares) or G2400A mutant (circles) ribosomes. The data are presented as mean values of three independent experiments; error bars show standard deviation. d SOL and TEL protect A2400 and A2401 of the 25S rRNA from chemical modification. The gel shows the primer extension analysis the 25S rRNA extracted from the untreated or dimethyl sulfate (DMS)-modified G2400A 80S ribosomes incubated without or with the antibiotics. The position of the cDNA bands corresponding to the nucleotide residues A2400 and A2401 are indicated by the arrows. The chemical modification of wt ribosomes in the presence of SOL and TEL and that of mutant ribosomes in the presence of ERY and azithromycin are shown in Supplementary Fig. 2b, c, respectively. The uncropped gel can be found in the Source data file. The result is a typical representative of two independent experiments.