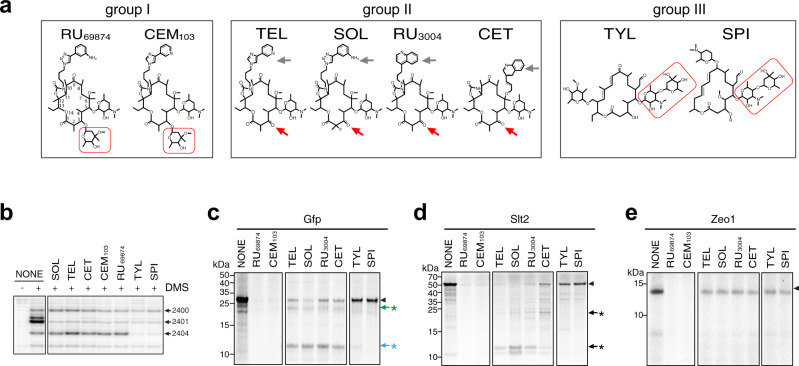
Fig. 6. The structure of the macrolide affects selective inhibition of translation.
a Structures of representative macrolide antibiotics grouped according to their chemical features: group I, 14-atom macrolactone ring carrying C3 cladinose sugar (framed in red); group II, 14-atom macrolactone ring with C3 keto group (red arrow) and extended alkyl-aryl side chain (gray arrow); group III, 16-member macrolactone with C5 disaccharide chain (framed in red). b Chemically different macrolides readily bind to the yeast G2400A mutant ribosome as revealed by the protection of 25S rRNA residues A2400(2058), A2401(2059) and, in the case of the group III compounds, A2404(2602) from DMS modification. The drugs were present at 100 µM. The primer extension products were resolved in the denaturing 6% polyacrylamide gel. Control samples (“NONE”) contained no macrolides. c–e SDS-gel analysis of the [35S]-labeled protein products accumulated during translation in the yeast cell-free system of templates encoding the Gfp reporter (c) or the Slt2 (d), and Zeo1 (e) yeast proteins. Translation reactions were supplemented with 100 µM of the indicated macrolides. The control reaction (NONE) contained no antibiotic. Arrowheads and arrows/asterisk represent the bands corresponding to the full-length and truncated products, respectively. The uncropped gel can be found in the Source data file. b–e Representative gels of two independent experiments.