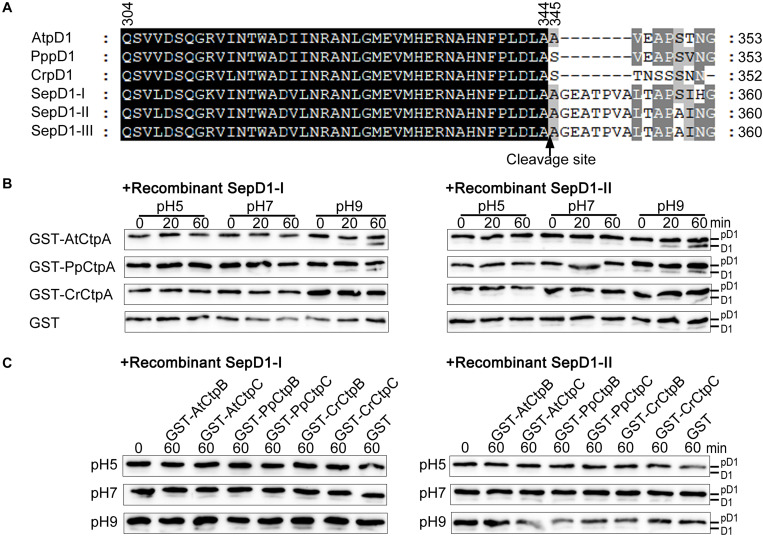
FIGURE 6.
Protease activity of eukaryotic Ctp on prokaryotic pD1. (A) C-terminal sequence comparison of pD1 from four evolutionary lineages of oxygenic photosynthetic organisms. AtpD1, A. thaliana pD1; PppD1, P. patens pD1; CrepD1, C. reinhardtii pD1; SepD1-I, -II, and -III, S. elongatus PCC 7942 pD1-I, -II, and -III. The numbers indicate the positions of the corresponding amino acids in pD1. The C-terminal cleavage site between amino acids 344 and 345 is indicated by a vertical arrow. (B) In vitro protease activity assays of eukaryotic CtpA using as substrate recombinant SepD1-I (left) and SepD1-II (right), which were produced by fusing GST with the C-terminal 57 aa fragment of SepD1-I or SepD1-II. The experiments were conducted as described in the legend of Figure 4. (C) In vitro protease activity assays of eukaryotic CtpB and CtpC using as substrate recombinant SepD1-I (left) and SepD1-II (right).