Abstract
Alzheimer’s disease (AD) is the most common cause of dementia, afflicting more than 5 million Americans; it is the 6th leading cause of death in the United States. As the population ages, the number of Americans with AD expected to increase dramatically. Understanding the cellular processes that lead to AD pathology is critical to designing meaningful therapeutic interventions. AD is characterized by the accumulation of aggregated proteins that may be toxic and may represent failure of the cell to normally process these proteins. One key to understanding sporadic AD lies in the genetics of families with highly penetrant histories of the disease. Mutations in subunits of a cellular trafficking complex known as COPI were found in families with AD and no other known AD-associated mutations. The COPI complex is involved in protein processing and trafficking within the cell. Intriguingly, several recent publications have found that components of the COPI complex can affect the metabolism of pathogenic AD proteins. We report here that reducing levels of the COPI subunit α-COP alters maturation and cleavage of amyloid precursor protein (APP), resulting in decreased release of Aβ−42 and decreased accumulation of the APP intracellular C-terminal domain. We also found that depletion of α-COP reduces uptake of proteopathic Tau seeds and reduces intracellular Tau self-association. Expression of AD-associated mutations in α-COP altered APP processing, resulting in increased release of Aβ−42 and increased intracellular Tau aggregation and release of Tau oligomers. Taken together, these results show that COPI coatomer function modulates the processing of both APP and Tau and that expression of AD-associated α-COP mutant proteins confers a toxic gain of function, which results in potentially pathogenic changes in both APP and Tau.
Keywords: Golgi apparatus, COPI coatomer, Amyloid precursor protein, Tau
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease and the most common cause of dementia in aging. The pathologic hallmarks of the AD brain are extracellular amyloid plaques consisting of Aβ (Selkoe and Hardy, 2016) and the intracellular neurofibrillary tangles (NFTs) composed of Tau (Sun et al., 2017). The cellular pathways that regulate the processing of proteins involved in these pathologic pathways could be the keys to unlocking the treatment for this devastating disease. A great deal has been learned about the AD state by the study of genetic variants implicated in familial forms of the disease. We report here a study of the role of COPI coatomer in the processing and trafficking of amyloid precursor protein (APP), prompted by previous studies showing that deletion of the COPI subunit Arcn1/δ-COP in cells reduced the release of Aβ from cells expressing the neuronal splice form of APP (Bettayeb et al., 2016a).
α-COP is a member of the heptameric COPI coatomer complex, involved primarily in Golgi-ER retrograde transport, endosomal trafficking, and macroautophagy (Beck et al., 2009b). Previously, we have shown that α-COP is involved in the transport of survival motor neuron (SMN) protein, loss of which causes the hereditary neuromuscular disease spinal muscular atrophy (SMA) (Peter et al., 2011). In cell culture and animal models of SMA, SMN mutants that are unable to bind α-COP cannot rescue neurite outgrowth defects that result from SMN knockdown (Custer et al., 2013). Overexpression of α-COP rescues SMN depleted cells and zebrafish, whereas α-COP point mutants that no longer bind SMN do not (Li et al., 2015).
Recent evidence has indicated that the COPI coatomer function regulates the processing of APP, which is cleaved in the cell to generate the toxic, aggregate-prone Aβ fragments. Proteomic analyses demonstrated that COPI coatomer members α-COP and β-COP interact with multiple isoforms of APP, and β-COP can be immunoprecipitated by APP only in the presence of an intact COPI coatomer (Andrew et al., 2019; Selivanova et al., 2006). Depletion of 2 subunits of the COPI coatomer increased retention of APP in the Golgi apparatus and decreased transport of APP to the cell surface (Bettayeb et al., 2016a; Selivanova et al., 2006). This resulted in decreased production of Aβ fragments. Genetic crosses between AD model mice and the Nur17 mouse, which harbors a mutation in δ-COP/Arcn1 that partially disrupts COPI-dependent intracellular trafficking (Xu et al., 2010), resulted in reduced Aβ levels and amyloid plaque burden and improved performance in memory tasks (Bettayeb et al., 2016b). Involvement of α-COP in AD pathogenesis is implied by genetic studies. Cluster analysis of microarray data from entorhinal cortex in mid-stage AD brains showed increased expression of α-COP in neurons with high degrees of pathology compared with unaffected cells in the same brain region (Guttula et al., 2012). Given all these findings, we set out to explore the role of α-COP in the processing APP. In addition, we explore for the first time the role of the COPI coatomer in Tau-driven pathology in AD.
2. Results
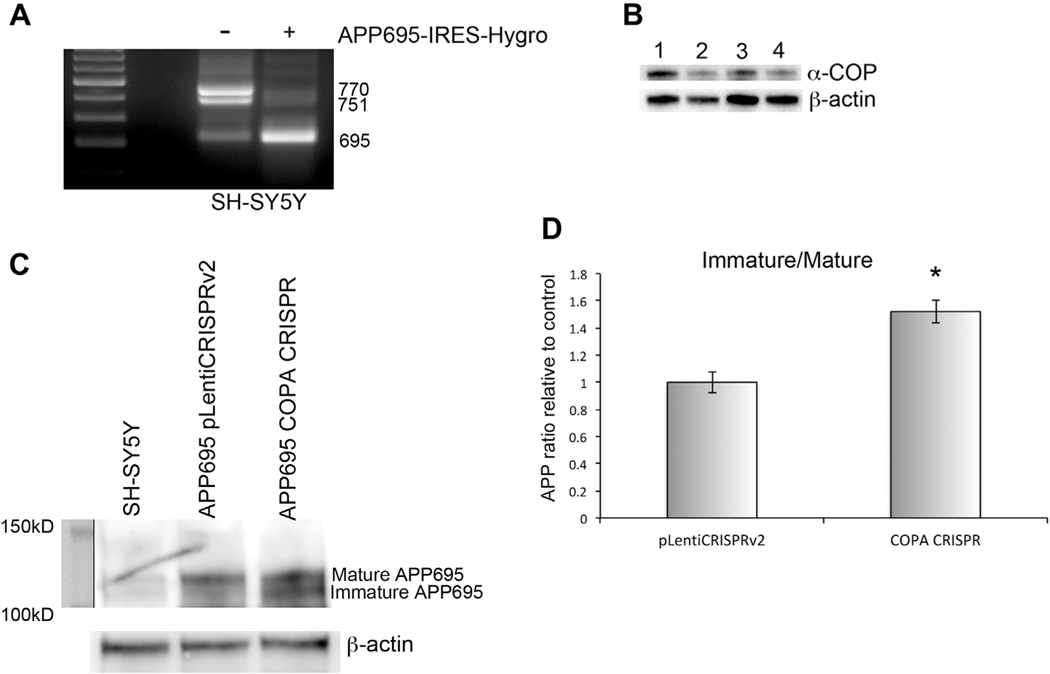
The COPI coatomer was originally implicated in the processing of APP by Selivanova et al. (2006). They described changes in APP delivery to the plasma membrane (PM) and its cleavage by gamma-secretase after depletion of the epsilon-subunit of the COPI complex. This work was performed in ldlF-2 cells, a clone of Chinese Hamster Ovary (CHO) cells with a temperature-sensitive point mutation in ε-COP, which renders the protein unstable, and results in extremely low levels of ε-COP at the restrictive temperature (Guo et al., 1996). We saw this work as a starting off point but wanted to move into a more neuronal context so that the APP processing enzymes such as the neuron-specific β-secretase BACE1 would be present. We chose the human neuroblastoma line SH-SY5Y. We generated a line of SH-SY5Y cells stably expressing the neuronal APP isoform, which can be cleaved to release Aβ−42 (APP695) using a previously published expression vector (Belyaev et al., 2010). Fig. 1A shows that the APP695-IRES-Hygro cells express almost exclusively the neuronal 695 amino acid APP isoform compared with the parental SH-SY5Y cells, which expressed primarily the 751 and 770 isoforms. The APP695-IRES-Hygro cells were used for subsequent experiments evaluating the processing of APP. To reduce the levels of α-COP in the cells, these were infected with a CRISPR Cas9 lentivirus targeting COPA. Although no bi-allelic knockout clones were detected, multiple haploinsufficient clones were isolated, reducing the levels of α-COP protein by approximately 50%. This recapitulates previous findings that only haploinsufficient lines could be generated with CRISPR guides targeting COPD/ARCN1 (Izumi et al., 2016). Fig. 1B shows a representative Western blot of the levels of α-COP in 3 clones relative to the control line expressing the control plasmid.
Fig. 1.
Depletion of α-COP alters processing of APP in SH-SY5Y cells. (A) RT-PCR using primers anchored in Exon 6 and Exon 9 amplifies 3 splice isoforms of human APP. Cells stably expressing APP695-IRES-Hygro express predominantly the 695 isoform (bottom band). (B) Western blot shows decreased levels of α-COP protein in the COPA CRISPR clones (lanes 2ò4) compared with controls. (C) Western blotting using the 22C11 antibody detects an increase in immature APP in COPA CRISPR clone compared with controls. (D) Quantitative densitometry from 3 separate Western blots shows a statistically significant increase in the ratio of immature/mature APP in COPA CRISPR cells compared with control. Error bars show standard error of the mean, and asterisk denotes statistical significance p < 0.05.
2.1. Reduction of α-COP alters processing of APP
To study the impact of α-COP depletion on the maturation of full-length APP695, we examined the levels of immature (N-glycosylated) and mature (N and O-glycosylated, tyrosyl-sulfated) APP protein by Western blot. Fig. 1C shows a representative Western blot demonstrating increased immature APP in COPA CRISPR clones compared with controls. Fig. 1D represents the combined results of 3 separate Western blot quantifications demonstrating a statistically significant shift in the ratio of immature to mature APP695 in cells with reduced α-COP compared with controls (p < .05 by Student’s t-test). This increase in immature APP695 after α-COP depletion agrees with previously published results, showing that dissociation of the COPI coatomer with Brefeldin A increased immature FLAG-tagged APP695 in murine N2a neuroblastoma cells (Saito et al., 2011).
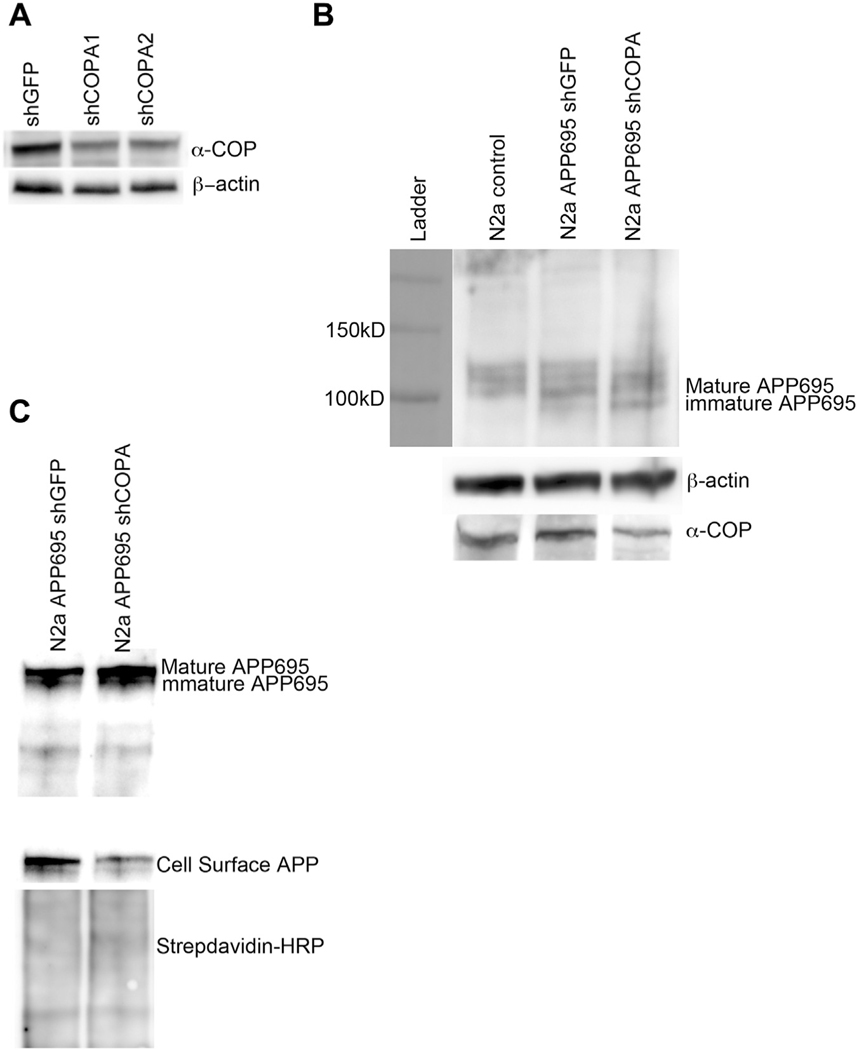
Selivanova et al. reported that in IdlF CHO cells cultured at restrictive temperatures to reduce the levels of ε-COP, there was decreased APP695 present at the PM (Selivanova et al., 2006). We attempted to replicate this finding in SH-SY5Y cells but found very low levels of APP were detected at the PM after labeling with cell impermeant biotin (data not shown). We chose to move to the murine neuroblastoma cell line N2a. Experiments in this cell line following stable knockdown of another COPI subunit, δ-COP/Arcn1, resulted in decreased delivery of APP695 to the PM. We used the same APP695-IRES-Hygro construct described previously to generate a line of N2a cells that stably expressed the neuronal isoform of APP. We then used previously validated shRNA against murine COPA (Li et al., 2015) to generate stable reduction of α-COP protein in this cell line. Stable expression of shRNA against green fluorescent protein (shGFP) served as a control N2a cell line. Fig. 2A shows a representative Western blot analysis of α-COP protein levels in the stable cell lines. We continued with shCOPA2, as this clone had the most consistent reduction of α-COP at the protein level. Fig. 2B shows that in N2a cells, we again found that reducing α-COP resulted in increased levels of the immature APP695 compared with the shGFP control line. Using cell impermeant biotin to label proteins resident at the PM followed by isolation with streptavidin beads, a human-specific antibody (E8B3O) detected decreased APP695 at the PM in the α-COP knockdown cells (Fig. 2C).
Fig. 2.
Depletion of α-COP alters processing of APP in N2a cells. (A) Representative Western blot from whole cell N2a lysates after puromycin selection and clonal isolation. Blotting with H3 antibody shows decreased levels of α-COP protein compared with β-actin loading controls. (B) Representative Western blot of whole cell lysate blotted with antibody 22C11 to visualize mature and immature forms of APP695 shows increased levels of immature APP695 in cells with stable knockdown of COPA. (c) Blotting of whole cell lysates and streptavidin-isolated proteins cell surface protein with human-specific APP antibody (E8B3O) shows decreased APP695 at the plasma membrane in COPA knockdown cells compared with controls. Blotting with HRP-conjugated Streptavidin confirms even loading of the cell surface proteins.
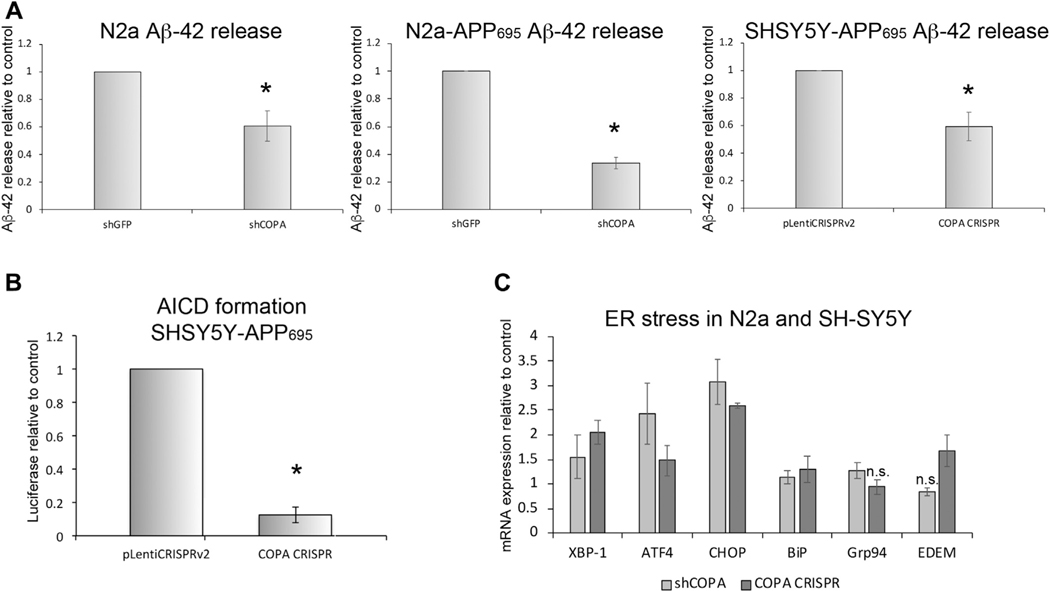
In addition to traveling through the secretory pathway to mature and reach the PM, APP is processed to yield a number of cleavage products. Cleavage by BACE-1 yields the C99 fragment, which can be further cleaved to produce Aβ−42. When released into the extracellular environment, Aβ−42 becomes the main component of the amyloid plaques, which are a pathologic hallmark of the Alzheimer’s brain (reviewed in a study by Esler and Wolfe, 2001). In N2a cells where δ-COP/Arcn1 was reduced by transfection with small interfering RNA, altered APP processing resulted in reduced release of Aβ−40 (Bettayeb et al., 2016a). The authors reported this finding as potentially protective against AD pathology despite the fact that many consider Aβ−42 to be the more pathologic APP cleavage product (Phillips, 2019). Therefore, we chose to measure the release of Aβ−42. We cultured both SH-SY5Y and N2a cells that had been selected to stably express APP695 along with reduced levels of α-COP and used ELISA to measure the accumulation of Aβ−42 in the cell culture supernatant, normalizing the levels to total protein in the cell lysates to control for differences in plating density. Fig. 3A shows that in both SH-SY5Y and N2a cells, after decreases in the level of α-COP either by CRISPR-mediated haploinsufficiency (SH-SY5Y) or by stable expression of shRNA (N2a), the release of Aβ−42 was reduced whether we measured murine Aβ−42 or total (human and mouse combined).
Fig. 3.
Depletion of α-COP reduces the release of APP fragments. (A) Combined results of ELISA measurements of murine Aβ−42 in N2a cells show decreased Aβ−42 in cell culture media from a clonal line with stable expression of shRNA against COPA (shCOPA) compared with a control clone expressing shRNA against GFP (shGFP). Similar results were detected from a clonal line of N2a cells expressing stably expressing human APP695 in addition to the shRNA. Combined results of ELISA measurements of Aβ−42 in SH-SY5Y cells show a similar significant decrease in Aβ−42 from cells where the level of α-COP protein has been reduced by CRISPR editing of the COPA gene. (B) This graph depicts the combined results of multiple luciferase assays in SH-SY5Y cells expressing a reporter plasmid to measure the formation of the AICD following cleavage of APP695 by γ-secretase. Reducing α-COP protein by CRISPR gene editing resulted in decreased luciferase signal compared with control cells expressing the pLentiCRISPRv2 with a nonspecific gRNA. (C) Quantitative RT-PCR shows that reducing the levels of α-COP in the presence of APP695 induces modest ER stress compared with controls. Error bars represent standard error of the mean, and asterisk denotes statistical significance p < 0.05. n.s. = not significant.
Recent research efforts have focused on the importance of the Amyloid Precursor Intracellular C-terminal Domain (AICD), a protein fragment which when derived from APP695, can be transcriptionally active (Belyaev et al., 2010; Cao and Sudhof, 2001; Multhaup et al., 2015). For an alternative method of measuring APP processing, we obtained SH-SY5Y cells that stably express APP695-Gal4 along with Gal4-UAS Luciferase to measure the production of the AICD (Cao and Sudhof, 2001; Zhang et al., 2007). After cleavage by BACE1 and γ-secretase, the AICD-Gal4 fragment translocates to the nucleus and activates transcription of luciferase. The COPA CRISPR lentivirus was again used to stably reduce the levels of α-COP protein in these cells, and 2 separate clones were selected. Fig. 3B shows that depletion of α-COP protein greatly reduces the levels of luciferase produced compared with lines expressing the pLentiCRISPRv2 alone, indicating that there is reduced production of AICD when α-COP levels are low. These results are consistent with the findings that Selivanova et al. reported on depletion of ε-COP in IdlF CHO cells; however, those experiments used a cleavage assay, which expresses only the C-terminal 99 amino acids of APP fused to activate the luciferase. That system does not reflect the full sequence of APP processing required to generate the AICD, only the γ-secretase cleavage step (Karlstrom et al., 2002). Taken together, these results demonstrate that reducing the levels of α-COP alters the processing of APP at multiple levels by impacting its maturation, transport, and cleavage and resulting in reduced release of the toxic Aβ−42 cleavage product. We believe that these changes would be protective against amyloid-mediated AD pathogenesis, and in fact, when mice with reduced COPI function were crossed onto a model of amyloid-driven AD, there was decreased accumulation of amyloid plaques in the brains as well as improved performance on behavioral tasks (Bettayeb et al., 2016b).
Altering the maturation of APP to favor the immature form can result in the accumulation of the immature form in the endoplasmic reticulum (ER), which could induce ER stress (Cavieres et al., 2015). To measure potential induction of the ER stress response, we used quantitative RT-PCR to compare the expression of a number of markers of ER stress and ERAD induction in both N2a and SH-SY5Y expressing APP695 cells after depletion of α-COP by either shRNA or CRISPR gene editing. Fig. 3C shows that reduction of α-COP protein levels in the presence of APP695 does appear to induce a modest induction of ER stress transcripts (p < 0.05 by analysis of variance [ANOVA] with post-hoc Tukey’s t-test), although this may also be a result of reducing the overall COPI function, as depletion of COPI coatomer has been shown to induce ER stress on its own (Claerhout et al., 2012).
2.2. α-COP and tau pathology
A second pathogenic protein involved in AD is Tau, which regulates microtubule dynamics, axonal transport, and neuronal morphology by binding and stabilizing the microtubule structure (Morris et al., 2011). Hyperphosphorylated Tau can accumulate to form NFTs, and it is believed that hyperphosphorylated Tau no longer associates with microtubules and becomes prone to aggregation (Chong et al., 2018). The appearance of NFTs strongly correlates with the onset and progression of dementia (Alafuzoff et al., 1987; Arriagada et al., 1992; Knopman et al., 2013). Both defective autophagy and increased ER stress have been reported in cells after depletion of COPI subunits (Claerhout et al., 2012), and both these conditions can lead to accumulation of hyperphosphorylated Tau (Piras et al., 2016).
2.3. Depletion of α-COP reduces uptake of proteopathic tau
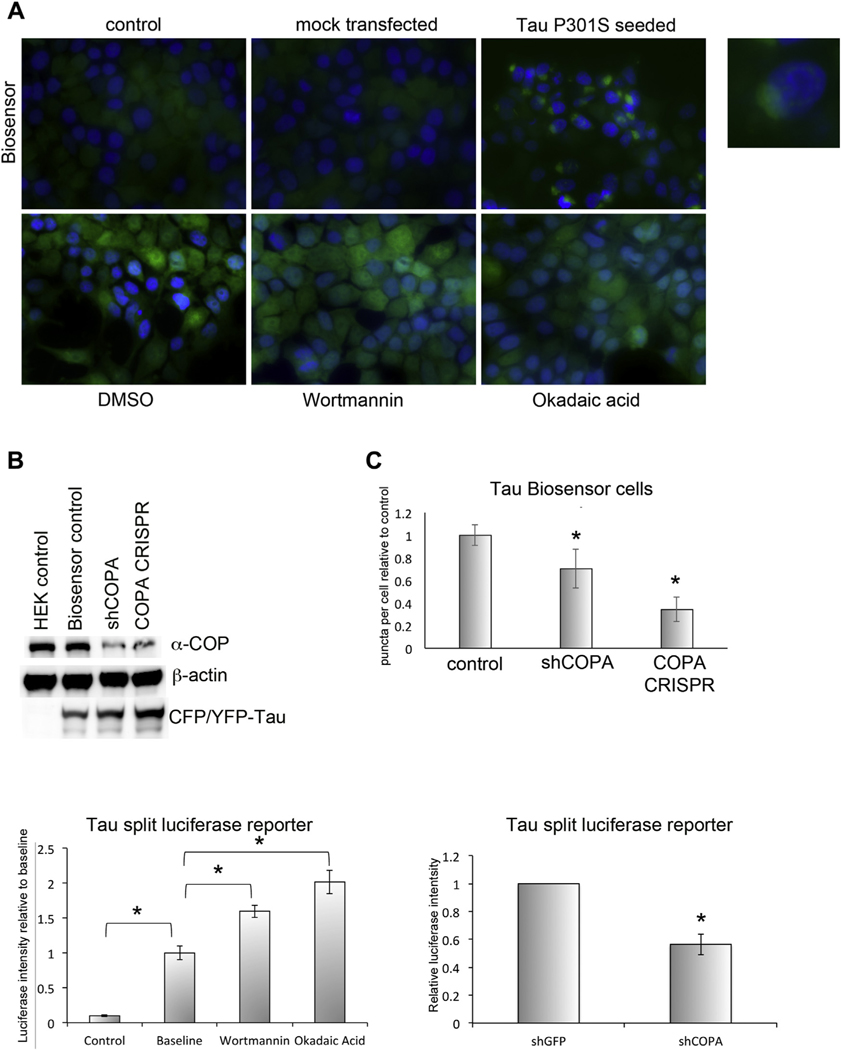
It has been reported in both animal and cell culture models that Tau may be able to seed pathology in neighboring cells in a prion-like fashion (Frost et al., 2009; Sanders et al., 2014). To study this phenomenon, we obtained the so-called Tau biosensor cells. Briefly, these HEK-293T cells express the Tau repeat domain with the pathogenic P301S mutation fused to either cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) (Furman et al., 2015). We reduced the levels of α-COP protein in these cells by selecting stable clones expressing either an shRNA against human α-COP or by infecting with LentiCrisprV2 and selecting the haploinsufficient clones. As mentioned previously, when using this same vector on SH-SY5Y cells, we were unable to isolate any COPA null cell lines. Fig. 4A shows representative micrographs of the biosensor cells. Under control conditions or mock transfection, these cells display minimal fluorescence in the green channel. However, transfection with proteopathic Tau seeds—in this case, brain exosomes derived from Tau P301S transgenic mice—induced FRET and a robust punctate GFP signal (Fig. 4A, inset). Fig. 4B shows that both the shRNA clone and the CRISPR clone have reduced the levels of α-COP protein without altering the levels of Tau-CFP/YFP. The cells were exposed to 5 μM Tau seeds and fixed 48 hours posttransfection. A blinded observer counted the number of GFP-positive cells per visual field from each condition for a total of approximately 200 cells. Fig. 4C shows the combined results of 3 separate transfections, and in each case, we found that reducing the levels of α-COP reduced the response of the cells to proteopathic seeding. Although these cells are extremely useful for the study of response to extracellular Tau seeds, we found that other treatments to induce Tau aggregation, such as okadaic acid or wortmannin, were unable to induce a FRET signal in these cells (Fig. 4A bottom row) and limited the utility of this cell line for further study of cell autonomous changes in Tau biology.
Fig. 4.
Depletion of α-COP alters Tau pathology in cells. (A) The Tau biosensor reporter line shows minimal fluorescence under control or mock transfection conditions, but when transfected with brain exosomes from symptomatic Tau P301S mice (5 μM), a strong FRET signal is produced, reflecting aggregation of the Tau-CFP and Tau-YFP proteins (inset). Treatment with wortmannin or okadaic acid increased background fluorescence comparably to the DMSO control (bottom row) and did not induce aggregation of the split- fluorescent Tau, indicating that Tau aggregation can only be induced by externally seeded Tau aggregates. (B) Western blot of whole cell lysate from Tau biosensor cells demon-strates that in cells stably expressing an shRNA against human COPA as well as in a second clone expressing the COPA CRISPR guide, α-COP protein levels are reduced compared with pLKO controls. Blotting with an antibody against GFP confirms that the split-fluorescent Tau is expressed at comparable levels in all clones. β-actin was used as a loading control. (C) When transfected with equal amounts of proteopathic Tau seeds, we find reduced FRET signal in cell lines with decreased levels of α-COP. (D) Co-transfection of N2a cells with Tau-Gluc1 and Tau-Gluc2 yields a roughly 10-fold increase in luciferase intensity compared with control cells transfected with Tau-Gluc1 alone. Treatment with either wortmannin or okadaic acid significantly increased the luciferase intensity produced by cells transfected with both Tau-Gluc1 and Tau-Gluc2 above baseline luciferase levels (p < 0.05 by Student’s t-test). (E) Transient transfection of stable N2a cell lines with the Tau split-luciferase reporter shows decreased luciferase signal after COPA knockdown compared with controls indicating a reduction in levels of Tau oligomerization. Error bars represent standard error of the mean, and asterisk denotes statistical significance p < 0.05.
2.4. Depletion of α-COP protects against intracellular tau aggregation
To adopt a more versatile reporter of pathogenic Tau changes, we obtained a Tau split-luciferase protein complementation assay vector, which takes advantage of the fact that proteins can be fused to 2 separate halves of Gaussia luciferase (Gluc) and on association can reconstitute luciferase activity (Remy and Michnick, 2006). The 2 Gaussia luciferase halves, Gluc1 and Gluc2, were fused to the C-terminus of human Tau 0NR4 isoform (Yan et al., 2016) (Nykanen et al., 2012). We chose to transfect the Tau-Gluc1 and Tau-Gluc2 into our stable N2a cell lines described previously in Fig. 2, expressing either a control shRNA against GFP or stably expressing shRNA against COPA. As a negative control, cells were transfected with Tau-Gluc1 alone. At baseline, we found a 10-fold increase in luciferase intensity when cells were transfected with both Tau-Gluc1 and Tau-Gluc2 compared with cells transfected with Tau-Gluc1 alone. As a positive control, cells transfected with Tau-Gluc1 and Tau-Gluc2 were treated with either wortmannin or okadaic acid to induce hyperphosphorylation of Tau, and both these treatments resulted in significantly increased luciferase intensity compared with baseline transfections (Fig. 4D) (Deng et al., 2005; Harris et al., 1993). Fig. 4E shows that knockdown of COPA reduces luciferase signal in cells transfected with Tau-Gluc 1 and 2, and similar results were found in HEK-293 cells in which α-COP protein levels were reduced by CRISPR knockout of COPA (not shown). Taken together, these results show that reduced protein levels of α-COP can protect against Tau-driven AD pathology both by decreasing the uptake of proteopathic Tau seeds and by reducing the levels of intracellular Tau aggregation. Combined with the apparent protective changes in APP trafficking and cleavage, demonstrating that modulating the COPI coatomer may be able to yield a multipronged protection, which impacts both amyloid and Tau-driven pathologic changes at the cellular level.
2.5. Mutations in α-COP associated with increased risk of AD
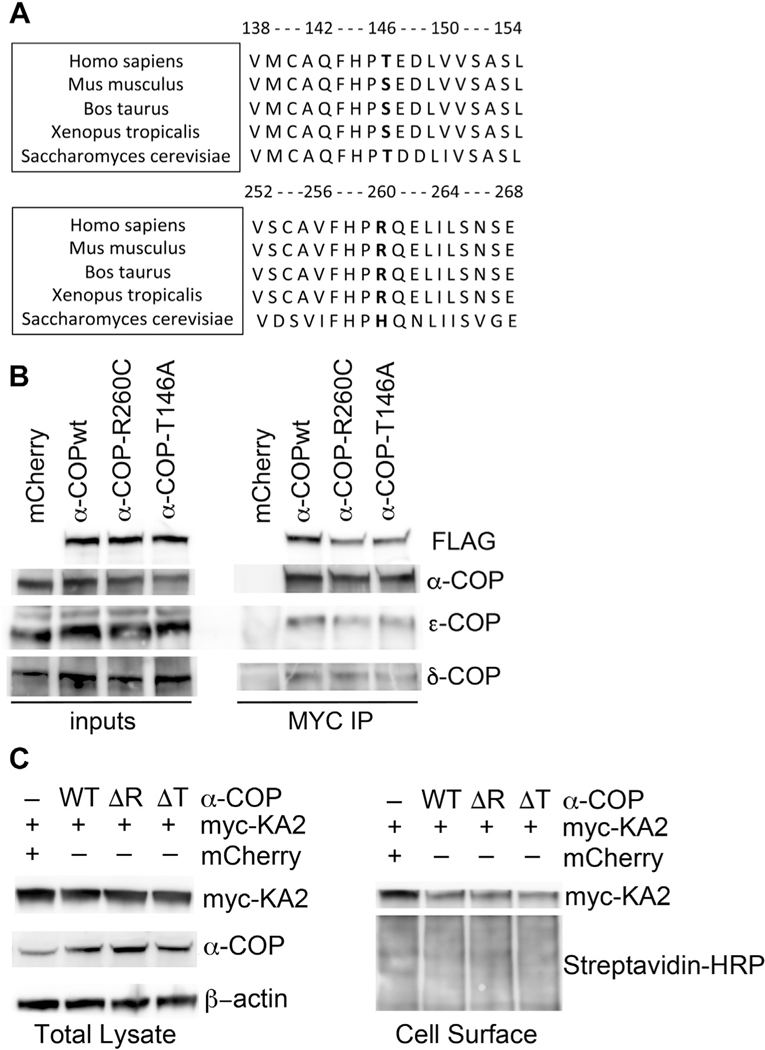
A great deal has been learned about the AD disease state by the study of genetic variants implicated in familial forms of the disease. Recent studies identified potential genetic links between the expression of the COPA gene and risk of AD, including a cluster analysis of microarray data (Guttula et al., 2012) and dynamic regulatory network reconstruction demonstrated altered COPA activity (Kong et al., 2014). Analysis of the National Institute of Mental Health Genetics Initiative Alzheimer’s Disease Study uncovered a number of small nucleotide polymorphisms in genes for COPI subunits associated with an increased risk of AD. Of particular interest were rare and highly penetrant variants in the coding region of COPA present in families where no other AD-associated mutations were detected. A mutation resulting in an amino acid change at position 146 from threonine to alanine (T146A) was detected in 20/29 affected carriers in 14 families. A second mutation resulting in an amino acid substitution at position 260 from arginine to cysteine (R260) mutation was present in 2/3 affected carriers in 1 family (Bettayeb et al., 2016b). The mutations lie in the highly conserved WD40 repeat domain (amino acids 1–285), which forms a β-propeller that appears to mediate the interaction of α-COP with COPI vesicle cargo (Eugster et al., 2000). Interestingly, T146 is conserved between humans and yeast but is a serine in other species (see alignment in Fig. 5A). T146 is predicted with a high degree of certainty to be phosphorylated (GPS 2.1), and the change to alanine, which cannot be phosphorylated, could result in significant structural changes. In humans, mutations in the WD40 repeats domain cause an autoimmune syndrome with exercise-induced dyskinesia, and the resulting mutant protein in this disorder appears to act as a dominant negative when overexpressed in Hela cells (Jensson et al., 2017; Watkin et al., 2015). The association of the T146A and R260C variants with increased risk of AD implies that these alter the function of α-COP in a way that is detrimental to neuronal cells.
Fig. 5.
AD-associated mutations in α-COP. (A) Sequence alignment shows conservation of T/S at amino acid position 146 and R at amino acid position 260 in a variety of organisms. (B) A representative Western blot of whole cell lysate (inputs) from cells transfected with either mCherry (control) or tagged human α-COP expression vectors (wild-type = WT) shows the expression of tagged α-COP vectors is approximately equal. Immunoprecipitation followed by Western blot analysis shows that both WT and AD-associated α-COP can co-immunoprecipitate members of the COPI complex. (C) Cells were transfected with myc-tagged kainate receptor (myc-KA2) in the presence of either an mCherry control or FLAG-tagged α-COP expression vectors. Wild-type, R260C (ΔR), and T146A (ΔT) all equally reduced the levels of myc-KA2 detected at the membrane.
To study the impact of these 2 mutations on the function of α-COP in relationship to AD pathology, the mutations were introduced into mammalian expression vector, including an MYC and FLAG tag at the C-terminus. Our first question was whether the α-COP mutant proteins could be stably expressed. In Fig. 5B, we show Western blot analysis of whole cell lysate from N2a cells expressing both wild-type and mutant α-COP. The input blots demonstrate that both T146A and R260C α-COP protein can be expressed at comparable levels to wild type (FLAG blot with FLAG-M2 antibody) but result in only a modest overexpression compared with endogenous levels α-COP blot with H3 antibody). Reports comparing AD brains to normal aging showed increased expression of COPI subunits, but we did not find that expression of R260C or T146A altered the levels of endogenous COPI members (Ciryam et al., 2016). Our second question was whether R260C and T146A α-COP would be able to enter the COPI coat. The COPI coatomer is a heptameric protein complex (Arakel and Schwappach, 2018). In the cytosol, COPI exists as 2 subcomplexes, α/β’/ε and δ/β/γ/ζ (Wang et al., 2016). To determine whether the T146A and R260C α-COP proteins enter a complete COPI heptamer, we used the MYC tag to immunoprecipitate the expressed α-COP and blotted back for COPI subunits from both halves of the subcomplexes, namely, ε-COP and δ-COP/Arcn1. Fig. 5B shows that both T146A and R260C α-COP coprecipitate members of each half of the COPI coatomer at comparable levels to wild-type α-COP, indicating that they are entering into a fully formed heptameric COPI complex. Their apparent ability to enter a fully heptameric COPI complex implies that the T146A and R260C α-COP may retain normal COPI function. To test this, we co-expressed either wild-type or T146A and R260C α-COP in the presence of a myc-tagged kainate receptor. COPI can retain proteins in the ER, as is the case for the kainite receptor subunit KA2. In the absence of other receptor subunits, KA2 is sequestered in the ER via interaction with COPI. Depletion of COPI components prevents this interaction and allows increased KA2 to reach the membrane (Vivithanaporn et al., 2006). We have previously used this assay to demonstrate that COPI coatomer function is decreased in cell models of SMA because of a reduction in COPI function (Custer et al., 2019). Fig. 5C shows that myc-KA2 is equally expressed when co-transfected with either an mCherry control or the various α-COP constructs. Western blot of biotinylated cell surface proteins followed by isolation with streptavidin beads shows that when co-expressed with mCherry, myc-KA2 escapes to the membrane, but in the presence of wild-type or T146A and R260C α-COP, COPI-dependent ER retention is used, reducing the levels of myc-KA2 detected in the cell surface fraction. This indicates that T146A and R260C α-COP are capable of carrying out normal COPI coatomer functions within the canonical secretory pathway.
2.6. AD-associated mutations in α-COP alter APP processing
As discussed previously, the intracellular trafficking and cleavage of APP is crucial to the generation of toxic Aβ species that accumulate in Alzheimer’s brains. As reported in Figs 1 and 2, we found that reducing levels of α-COP shifted the ratio of immature to mature APP in 2 neuron-like cells lines. There was no consistent alteration in the ratio between the immature and mature forms of APP695 when co-expressed with either T146A or R260C α-COP in SH-SY5Y, N2a, or HEK cells. Using N2a cells to examine the delivery of APP to the PM, we found that expression of the AD-associated α-COP mutant proteins consistently reduced the levels of human APP695 found at the PM (Fig. 6A). In this particular assay, it appears that these mutant proteins may phenocopy the α-COP knockdown conditions, but it is unclear whether the reduced levels of APP at the PM reflect a lack of delivery or rather an increased turnover in the presence of mutant α-COP proteins.
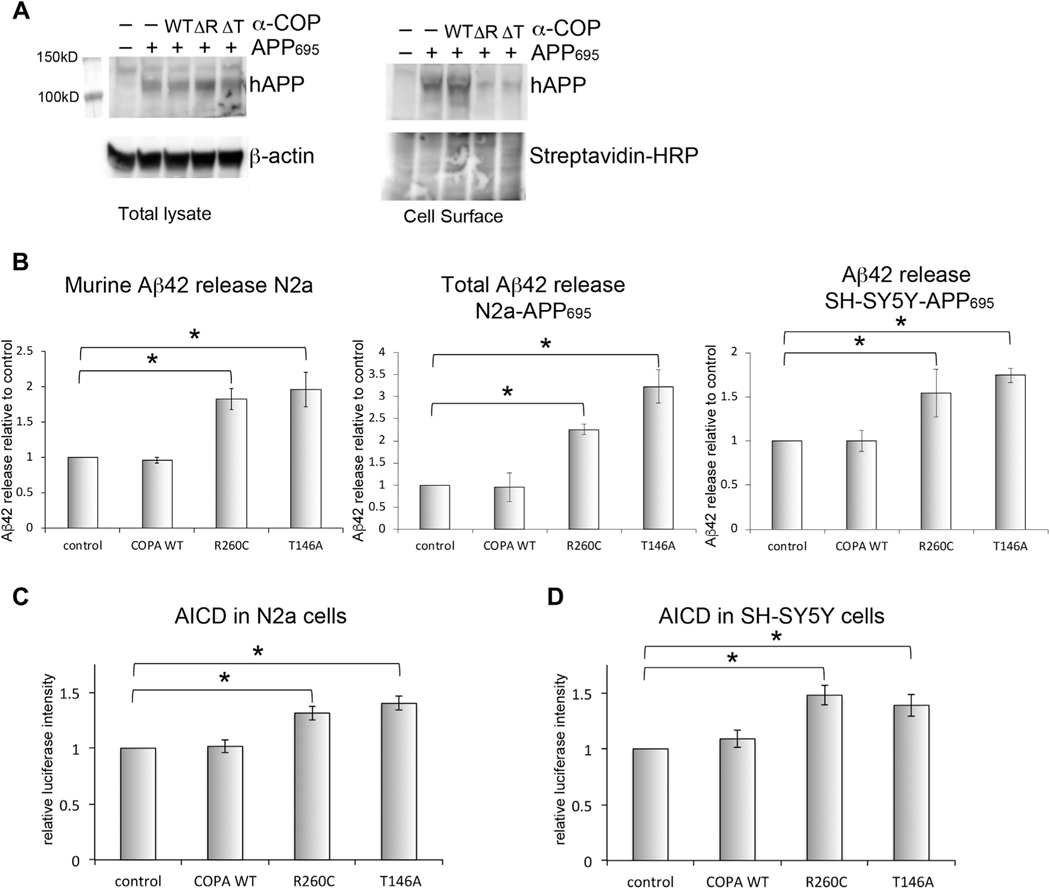
Fig. 6.
AD-associated mutations in α-COP affect APP processing. (A) Whole cell lysate from N2a cells expressing APP695 and α-COP vector probed with antibody E8B3O to visualize human APP695. When plasma membrane proteins were isolated by streptavidin pull-down after labeling with cell impermeant biotin, there was reduced plasma membrane–resident APP in the presence of R260C (ΔR) or T146A (ΔT) α-COP compared with control cells expressing APP695 alone or APP695 and α-COP wildtype (WT). The lower portion of the blot was probed with streptavidin-HRP as a loading control for biotinylated proteins. (B) ELISA quantification of Aβ−42 levels in cell culture supernatant shows increased release of Aβ−42 in the presence of R260C or T146A α-COP compared with wild-type or controls whether measuring murine Aβ−42 in N2a cells, combined Aβ−42 in N2a-APP695 or human Aβ−42 in SH-SY5Y cells. (D) α-COP expressing N2a cells transfected with APP-C99-GV and Gal4-luciferase show increased luciferase signal in the presence of R260C or T146A α-COP, indicating increased formation of AICD. (E) SH-SY5Y cells stably expressing a full-length APP γ-secretase reporter also show increased luciferase signal when expressing R260C or T146A α-COP, indicating increased formation of AICD. Error bars represent standard error of the mean. Asterisk denotes statistical significance p < 0.05.
To determine the impact of expressing AD-associated α-COP proteins on the release of Aβ−42, we measured the levels of Aβ−42 in the cell culture supernatant from N2a, N2a-APP695, and SH-SY5Y-APP695 cells expressing WT, R260C or T146A α-COP, using cells stably expressing pcDNA3 as the control. In both cell types expression of AD-associated mutant, α-COP proteins resulted in increased release of the toxic Aβ−42 species compared with expression of wild-type α-COP or control cultures (Fig. 6B). To examine APP cleavage inside the cell by γ-secretase, we used the SH-SY5Y cells described in Fig. 3. We also transfected the APP-C99-GV into a line of N2a cells stably expressing both Gal4-luciferase and α-COP. In each system, we found that expression of AD-associated α-COP mutants increased luciferase signal, indicating that the increase in the production of the AICD resulting from cleavage by γ-secretase (Fig. 6C and D). Taken together, these results demonstrate that AD-associated α-COP could promote amyloid-driven pathogenesis by altering the intracellular trafficking of APP to result in increased cleavage and release of amyloidogenic fragments.
2.7. AD-associated mutations in α-COP increase tau oligomerization and release
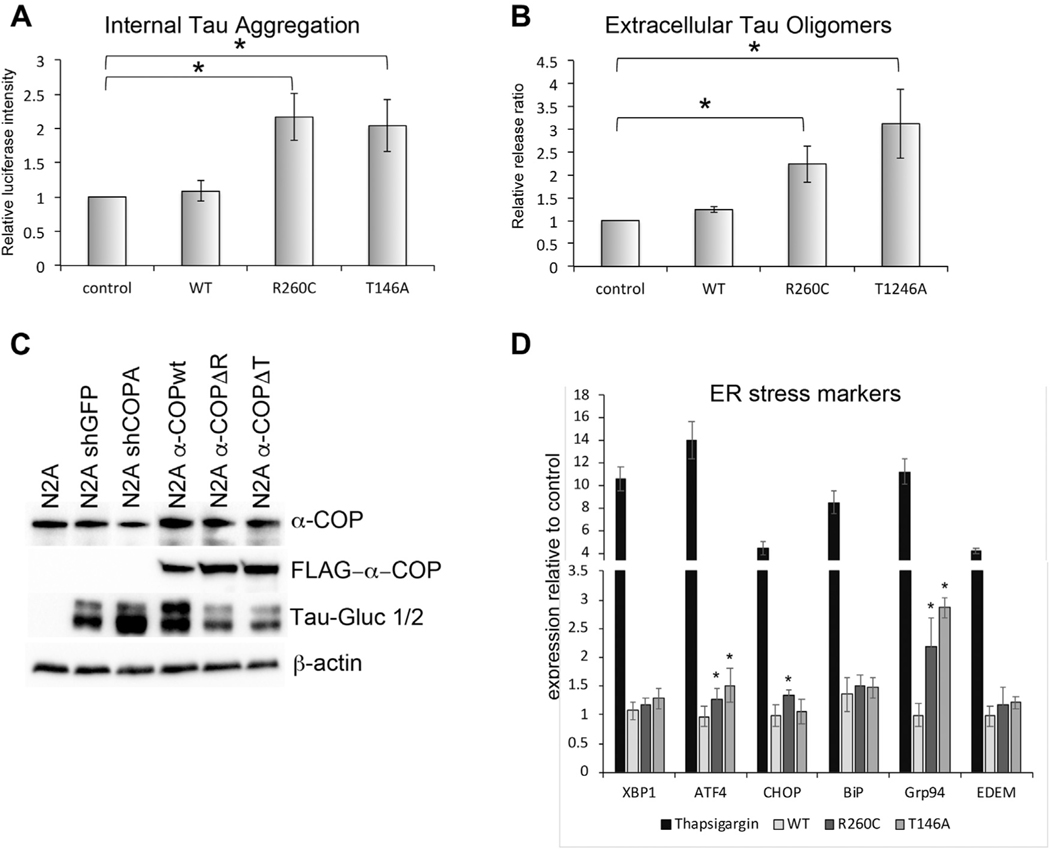
Given that reduction of α-COP appeared to be protective for cellular insults mediated by both amyloid and Tau, we sought to test the impact of expressing AD-associated α-COP mutant proteins on Tau aggregation. To address this, we used the split-luciferase system described in Fig. 4 in the highly transfectable N2a cells. The Tau-Gluc1 and Tau-Gluc2 constructs were transfected into N2a cells stably expressing wild-type, R260C and T146A α-COP, or a vector only control line of N2a cells. Fig. 7A shows that expression of R260C and T146A significantly increased Tau luciferase signal relative to control cells or cells expressing wild-type α-COP. It has been reported by a number of groups that dimerized and oligomerized Tau can be released by cells and taken up by neighboring cells to seed NFTs and propagate Tau pathology (Wang et al., 2017; Wegmann et al., 2016). To assess the impact of AD-associated α-COP mutations on Tau release, we also measured the luciferase activity in the cell culture supernatant and normalized the levels of luciferase in the media to the internal luciferase signal to calculate the fraction of aggregated Tau released in the various stable N2a lines. Fig. 7B shows that in clonal lines stably expressing either R260C or T146A α-COP, there is increased release of Tau compared with control lines or cells stable expressing wild-type α-COP. These data demonstrate that not only do the AD-associated α-COP mutants promote Tau aggregation intracellularly, which we predict would increase the formation of NFTs, these mutants also promote the release of proteopathic Tau that could infect neighboring cells in a prior-like fashion. Fig. 7C shows a Western blot of the stable N2a lines after transfection with the Tau split-luciferase reporter and confirms that the increases in luciferase activity seen in the presence of R260C or T146A α-COP is not the result of increased uptake or expression of the Tau-Gluc reporters. Although their ability to successfully retain myc-KA2 implies that the AD-associated α-COP mutants are capable of carrying out traditional COPI vesicle functions, their presence in the ER could still impede some aspects of COPI function, leading to the induction of an ER stress response. Several groups have reported increases in Tau hyperphosphorylation after ER stress (Ho et al., 2012; Liu et al., 2012). Using a series of RT-PCR primers to measure the levels of ER stress–induced transcripts (Oslowski and Urano, 2011), we examined the N2a cells stably expressing AD-associated α-COP vectors compared with wild-type or controls. As a positive control, we treated cells with thapsigargin, which induced a rapid increase in the ER stress indicators compared with controls. In contrast, expression of AD-associated α-COP induced very little indication of ER stress. We did observe increased expression of ATF4 and Grp94 compared with control cultures, and expression of R260C α-COP significantly increased expression of CHOP, but it was not statistically different from cells expressing wild-type or T146A α-COP (ANOVA with post-hoc Tukey’s t-test). These increases in expression of a subset of ER stress–induced transcripts suggest that some of the increase in intracellular Tau could be a result of increased ER stress, but the induction appears minimal compared with the robust induction seen in response to thapsigargin. Combined, these results implicate 2 previously uncharacterized α-COP mutations in the processing of both APP and Tau and provide a link between the Golgi apparatus, COPI coatomer function, and both axes of pathogenesis in AD.
Fig. 7.
AD-associated mutations in α-COP promote pathologic changes in Tau. (A) N2a cells expressing R260C or T146A α-COP show increased luciferase signal relative to controls when transfected with a split-luciferase Tau reporter, indicating an increase in Tau aggregation. (B) Expression of R260C or T146A α-COP in N2a cells results in an increased ratio of external aggregated Tau to internal aggregated Tau as expressed by the ratio of cell supernatant luciferase to whole cell lysate luciferase, indicating that cells expressing AD-associated α-COP mutations release increased amounts of potentially proteopathic aggregated Tau. (C) Western blot analysis of N2a cells stably expressing shRNA against either GFP or COPA or stably expressing α-COP (wildtype, R260C or T146A). Cells were transfected with Tau-Gluc 1 and Tau-Gluc2, and the blot was probed with antibody against human Tau. Expression of the split Tau reporter is roughly equal, and differences do not account for the increased luciferase activity seen when the reporter was expressed in the presence of R260C or T146A α-COP. (D) Quantitative RT-PCR was used to measure indicators of ER stress, using thapsigargin treatment as a positive control. Expression of R260C and T146A α-COP caused significant increases in ATF4 and Grp94 compared with cells expressing wild-type α-COP or mCherry controls. Error bars represent standard error of the mean. Asterisk denotes statistical significance p < 0.05.
3. Discussion
We find that in neuron-like cell lines, low levels of α-COP, a member of the COPI coatomer, altered the processing of APP, resulting in decreased Aβ−42 release and decreased AICD formation. We would predict that in the AD brain, these changes would be protective against amyloid-driven pathology. These findings are consistent with reports in cell culture and animal models of AD with reduced levels of Arcn1/δ-COP, where the investigators reported decreased release of APP fragments and decreased amyloid pathology in the brains of double-transgenic mice (Bettayeb et al., 2016a,b). In humans, 2 hereditary diseases have been linked to mutations in COPI coatomer members, in which amino acid changes result in deleterious developmental effects (Izumi et al., 2016; Jensson et al., 2017; Patwardhan and Spencer, 2019; Watkin et al., 2015). However, because AD is a disease of aging, it may be that a reduction in COPI function in the adult brain would be tolerated and allow this to be a strategy route for meaningful therapeutic intervention. A recent report analyzing proteomic changes that took place in response to oligomeric Aβ−42 exposure found that in cells carrying the so-called London mutation (APP V717I), which causes one of the most common forms of familial AD, COPI subunits were downregulated (Goate et al., 1991; Sackmann and Hallbeck, 2020). We hypothesize that this may be a protective mechanism to reduce amyloidogenic processing of APP in the face of Aβ−42 pathology in the microenvironment. A study of newly synthesized proteins in hAPP/PS1 mice found that ε-COP synthesis increased in the hippocampus in the presymptomatic period, whereas γ2-COP decreased in the cortex, indicating that the individual COPI subunits may have distinct responses and functions over the course of amyloid-driven pathology (Ma et al., 2020). COPI vesicle biogenesis is controlled by Arf1, a small GTPase of the Ras superfamily, making the COPI coatomer a potential target for small-molecule GTPase inhibitors as a therapeutic intervention in AD (Beck et al., 2009a; Cromm et al., 2015).
This work is the first to examine the role of COPI coatomer in Tau uptake and aggregation, and we found a similarly protective impact of reducing the levels of α-COP protein to what we observed when examining APP processing. Reduced α-COP was reduced uptake of extracellular proteopathic Tau. In an AD brain, this could result in slower spread of Tau pathology to neighboring brain regions. Recent work demonstrates that in the HEK Biosensor cell line, uptake of proteopathic Tau seeds is primarily regulated by heparan sulfate proteoglycans (Stopschinski et al., 2020). Although COPI has not been directly implicated in heparan sulfate proteoglycan–mediated endocytosis, it was reported that COPI is required for recycling of exocytic SNARE proteins at the PM (Xu et al., 2017), as well as entry of arginine-rich peptides and viral entry (Cureton et al., 2012; Tsumuraya and Matsushita, 2014), so reduction of α-COP may slow the dynamics of PM recycling and therefore protect the cells by slowing the uptake of proteopathic Tau seeds. In addition, COPI coatomer function is required for normal endosomal trafficking and retrieval of endosomal contents, so even if the initial endocytic event is not COPI regulated, the regulation of endocytosed products may be leading to an effective change in Tau uptake without directly influencing endocytosis (Daro et al.,1997; Gabriely et al., 2007; Tamayo et al., 2008). In both N2a and HEK-293Tcells with reduced levels of α-COP, we found that split-luciferase Tau was less likely to self-associate. These results are in apparent contrast to a previous report perform in HEK cells overexpressing aggregate-prone Tau where knockdown of Golgin 84 or treatment with Brefeldin A caused Golgi fragmentation and increased Tau hyperphosphorylation (Jiang et al., 2014). Although the secretion of proteopathic Tau was previously linked to Golgi fragmentation resulting from prolonged neuronal hyperactivity (Mohamed et al., 2017), using the Tau split-luciferase assay, we did not find that reducing the levels of α-COP significantly affected secretion of oligomerized Tau.
This report is the first to characterize AD-associated mutations in α-COP, and recent reports found significant increased risk for AD conferred by small nucleotide polymorphisms in additional COPI subunits as well (Bettayeb et al., 2016b; Yang et al., 2019). We predicted that if the AD-associated amino acid changes in α-COP resulted in dominant-negative changes, the mutants would phenocopy a COPA knockdown. In our assays, we find that expression of the R260C and T146A α-COP variants result in opposing changes in APP and Tau dynamics compared with conditions of low α-COP, indicating that they may retain canonical COPI coatomer function. The idea that the R260C and T146A variants retain normal α-COP function was further supported by our finding that they appear to successfully complex with endogenous COPI coat members as evidenced by their ability to coprecipitate members from both halves of the COPI coatomer. As they do not phenocopy COPA knockdown in our assays of APP and Tau processing, we can infer that the R260C and T146A mutations confer a toxic gain of function that is not tolerated in neuronal cells. In future studies, it will be critically important to perform proteomic experiments to determine which novel interactions may be present to mediate this apparently increased toxicity.
Recently, a third pathogenic Alzheimer’s axis has been identified. Whole genome sequencing studies identified variants in Trem2 (Triggering Receptor Expressed on Myeloid cells 2) that increase the risk of developing AD (Jonsson et al., 2013), whereas other Trem2 mutations cause frontotemporal dementia (Le Ber et al., 2014) or Nasu-Hakola syndrome (Paloneva et al., 2002). Conversely, a missense mutation (S144G) may be protective against AD (Ulrich et al., 2017). A recent publication demonstrated that knockdown of δ-COP resulted in accumulation of mutant Trem2 in the ER-Golgi intermediate compartment (ERGIC) because of failure of COPI coatomer to return the protein to its normal steady-state ER localization (Sirkis et al., 2017). These data indicate that in addition to impacting the trafficking and processing of APP and Tau, modulating COPI coatomer function could also impact the processing and trafficking of Trem2, which could potentially be helpful or harmful depending on the point of AD pathogenesis.
4. Materials and methods
4.1. Cell culture
N2a, SH-SY5Y, and Biosensor cells were maintained in high- glucose DMEM with 10% fetal bovine serum and Normocin. All transfections were performed using Polyethylenimine (PEI) at a ratio of 1 μg DNA/2 μg PEI, and cells were transected overnight in serum-free DMEM.
4.2. Selection of stable APP695 cell lines and APP isoform expression analysis
An expression vector was obtained from Dr Nigel Hooper (University of Manchester), encoding the 695 amino acid isoform of human APP as well as a hygromycin resistance gene (APP695-IRES-Hygro). The vector was transfected into N2a and SH-SY5Y cells. Cells were transfected with PEI, and hygromycin was added 48 hours after transfection at a concentration of 100 μg/mL. After 10 days of selection, single clones were isolated, and APP695 expression was confirmed by Western blot with antibody 22C11 (EMD Millipore). For analysis of APP isoform expression in SH-SY5Y, total RNA was isolated from whole cells using the PureLink RNA Micro Kit (Invitrogen 12183016), then converted to complementary DNA using SuperScript-III (Invitrogen, 18080051). Primers were designed to amplify from exon 6 to exon 9 and were able to identify APP695, APP751, and APP770 (hAPP exon 6F 5″-AGG AAC CCT ACG AAG AAG CC-3″ hAPP exon 9R 5′-ATT CTC TCT CGG TGC TTG GC).
4.3. Selection of stable shRNA and CRISPR expression lines
For knockdown of murine COPA or GFP control, shRNA expression vectors were obtained from the Mission shRNA collection in the pLKO.1 vector backbone (SHC005 and TRCN0000313323, respectively). Forty-eight hours after transfection, transfected cells were selected by culturing in media containing puromycin (1–2 μg/mL), and colonies were isolated to generate clonal lines, which were screened by Western blot to identify lines with stable decrease in α-COP protein. CRISPR vector in the pLentiCRISPRv2 backbone targeting exon 5 of the human COPA gene was obtained from Genscript (gRNA sequence ATGATCAGACCATCCGAGTG). SH-SY5Y and HEK-293T Biosensor cells were transfected with either pLentiCRISPRv2 with a scrambled gRNA (GTATTACTGATATTGGTGGG). Forty-eight hours after transfection, transfected cells were selected by culturing in media containing puromycin (1–2 μg/mL), and colonies were isolated to generate clonal lines, which were screened by Western blot to identify lines with stable decrease in α-COP protein.
4.4. APP expression, trafficking, and cleavage
For visualization of mature and immature forms of full-length APP, cells were lysed for 20 minutes on ice in 150 mM NaCl, 50 mM Tris, and 1% Triton-X and clarified by centrifugation at 16,000 rpm. Protein concentration of the clarified lysate was determined using a BCA assay (Pierce BCA Kit, catalog # 23,225). Lysates were then added to 2X Laemmli buffer and separated by SDS page on 4%–12% gradient Bis-Tris gels and transferred to PVDF membrane. After blocking in TPST with 5% milk, the membranes were probed overnight with antibody 22C11 (EMD Millipore MAB348, 1:1000) or antibody E8B3O for the detection of human APP in mouse cells (Cell Signaling Technologies, #29765, 1:1000). The ratio of immature to mature APP was calculated by gel densitometry using ImageJ on 3 separate blots, and error bars show the standard error of the mean. To determine the amount of APP present at the cell membrane, PM proteins were labeled with cell impermeant biotin using Sulfo-NHS-SS-Biotin as previously described (Huang, 2012). After isolation of biotinylated proteins with streptavidin beads, beads were boiled in 2X Laemmli, and Western blotting was performed as described previously. Aβ−42 levels were measured by ELISA using the LEGEND MAX ELISA kit (BioLegend #842401) to detect human and combined Aβ−42 or the Amyloid beta 42 Mouse ELISA kit (Invitrogen #KMB3441) for the detection of murine Aβ−42 in N2a cells. For the detection of internal Aβ−42, cells were lysed in 150 mM NaCl, 50 mM Tris with 1% Triton-X and diluted 1:5 before use. Aβ−42 concentrations were normalized to total cell lysate protein to control for differences in plating density. SH-SY5Y cells stably expressing full-length human APP695-GV and Gal4-luciferase were obtained from Dr Saunders (Temple University). Stable lines expressing COPA CRISPR and control vectors as well as AD-associated COPA mutants were selected as described previously. Once knockdown of α-COP and expression of mutant α-COP proteins were confirmed, the luciferase assay was performed using the Luciferase Assay System (Promega, E1500). Cells were plated in 7 cm tissue culture dishes and lysed in 700 μL Cell Lysis Buffer (CLB, provided with kit). For assays measuring gamma-secretase cleavage using the C99-GV vector, cells were transiently transfected using PEI with the C99-GV, the Gal4-luciferase reporter, and pRenilla. Forty-eight hours after transfection, luciferase activity was measured using the Promega Dual-Glo assay kit as directed (Promega, #E2920) and normalized to renilla.
4.5. Tau proteopathic seeding
Biosensor HEK-293T cells were obtained from Dr Cristian Lasagna-Reeves. Cells were transfected with either shRNA against human COPA or COPA CRISPR, and clones with reduced α-COP protein expression were selected with puromycin. The source of Tau see material was brain homogenate mouse brain lysate from transgenic mice overexpressing human Tau (PS19 P301S) in TBS (without detergent) diluted to a final protein concentration of 6.6 μg/μL. Cells were plated on collagen-coated coverglass for 24 hours, then exposed to brain lysate combined with Lipofectamine 2000 at 1 μg protein/mL for 24 hours. Cells were incubated in fresh media for an additional 24 hours, then rinsed with phosphate-buffered saline and fixed for 5 minutes in 4% PFA before mounting with Prolong Gold (with DAPI). Ten random fields were captured for each condition, and a blinded observer counted the percentage of cells with GFP-positive puncta per image. This experiment was performed 3 times.
4.6. Tau oligomerization
Split-luciferase Tau vectors were provided by Dr Henri Huttunen, the University of Helsinki. Stable cell lines described previously (N2a and SH-SY5Y) were plated in 7 cm dishes and transiently transfected overnight with 3.5 μg Tau-Gluc1 and Tau-Gluc2 using PEI. After the overnight transfection, the media were changed to high-glucose DMEM with no Phenol Red (Gibco 21063029). Forty-eight hours after media change, the cells were lysed in 700 μL PLB, and luciferase activity was determined using the Dual-Luciferase assay kit (E1910). The same kit was used to determine luciferase activity in the cell culture supernatant. Media were harvested and spun at 1000 rpm for 5 minutes, then equal amounts of media and Passive Lysis Buffer (provided) were combined, and luciferase activity determined as recommended by the product insert. The ratio of cell culture media luciferase intensity was divided by the luciferase signal in the whole cell lysate to calculate the percent Tau released. We note that although the split luciferase in this assay reporter is Gaussia luciferase and should therefore be compatible with kits that provide coelenterazine as the substrate, we were only able to observe consistent luciferase activity using the Promega Dual-Luciferase kits with both the luciferase assay buffer and the stop and glow substrate present.
4.7. Measuring ER stress
To induce ER stress, N2a cells were treated with Thapsigargin (100 nM for 6 hours) as a positive control. Total RNA was isolated using TRIzol reagent and the PureLink Micro kit (Invitrogen, #12183016) followed by conversion to complementary DNA using the iScript system (Biorad #1708840). Quantitative RT-PCR was performed with iQ SYBR green supermix reagent (Biorad #1708880). Primers for transcripts reflecting ER stress induction were previously published (Oslowski and Urano, 2011), and β-actin was used as the control.
4.8. Kainate receptor trafficking assay
N2a cells were transfected with a previously characterized myc-tagged kainate receptor subunit (myc-KA2) combined with either mCherry or α-COP expression vectors (Vivithanaporn et al., 2006). Forty-eight hours posttransfection, cell surface proteins were labeled with cell-impermeant Sulfo-NHS-SS-Biotin as described previously. Biotinylated proteins were isolated with magnetic Streptavidin beads and eluted by boiling in 2x Laemli buffer. Total cell lysates and the biotinylated cell surface fraction were analyzed by Western blot with anti-Myc antibody (clone 9E10). HRP-conjugated streptavidin was used as a loading control for the cell surface fraction, and β-actin was used as a loading control for the whole cell lysates.
4.9. Statistical significance
For pair-wise comparisons, statistical significance was determined by Student’s t-test. For comparison of multiple groups, ANOVA was performed with post-hoc Tukey’s t-test. Statistical significance was determined as p < 0.05.
Acknowledgments
This work was supported by funding from the National Institutes of Neurological Disorders and Stroke (NINDS 1R01NS082284-01A1). L.A. was supported by the Stark Neuroscience Research Institute’s Medical Neuroscience Summer Research Program (SNRI MNSRP).
Footnotes
Disclosure statement
The authors have no actual or potential conflicts of interest.
References
- Alafuzoff I, Iqbal K, Friden H, Adolfsson R, Winblad B, 1987. Histopathological criteria for progressive dementia disorders: clinical-pathological correlation and classification by multivariate data analysis. Acta Neuropathol. 74, 209–225. [DOI] [PubMed] [Google Scholar]
- Andrew RJ, Fisher K, Heesom KJ, Kellett KAB, Hooper NM, 2019. Quantitative interaction proteomics reveals differences in the interactomes of amyloid precursor protein isoforms. J. Neurochem 149, 399–412. [DOI] [PubMed] [Google Scholar]
- Arakel EC, Schwappach B, 2018. Formation of COPI-coated vesicles at a glance. J. Cell Sci 131. [DOI] [PubMed] [Google Scholar]
- Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT, 1992. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology 42, 631–639. [DOI] [PubMed] [Google Scholar]
- Beck R, Adolf F, Weimer C, Bruegger B, Wieland FT, 2009a. ArfGAP1 activity and COPI vesicle biogenesis. Traffic 10, 307–315. [DOI] [PubMed] [Google Scholar]
- Beck R, Rawet M, Ravet M, Wieland FT, Cassel D, 2009b. The COPI system: molecular mechanisms and function. FEBS Lett. 583, 2701–2709. [DOI] [PubMed] [Google Scholar]
- Belyaev ND, Kellett KA, Beckett C, Makova NZ, Revett TJ, Nalivaeva NN, Hooper NM, Turner AJ, 2010. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J. Biol. Chem 285, 41443–41454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettayeb K, Chang JC, Luo W, Aryal S, Varotsis D, Randolph L, Netzer WJ, Greengard P, Flajolet M, 2016a. deltα-COP modulates Abeta peptide formation via retrograde trafficking of APP. Proc. Natl. Acad. Sci. U S A 113, 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettayeb K, Hooli BV, Parrado AR, Randolph L, Varotsis D, Aryal S, Gresack J, Tanzi RE, Greengard P, Flajolet M, 2016b. Relevance of the COPI complex for Alzheimer’s disease progression in vivo. Proc. Natl. Acad. Sci. U S A 113, 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Sudhof TC, 2001. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120. [DOI] [PubMed] [Google Scholar]
- Cavieres VA, González A, Muñoz VC, Yefi CP, Bustamante HA, Barraza RR, Tapia-Rojas C, Otth C, Barrera MJ, González C, Mardones GA, Inestrosa NC, Burgos PV, 2015. Tetrahydrohyperforin inhibits the proteolytic processing of amyloid precursor protein and enhances its degradation by Atg5-dependent autophagy. PLoS One 10, e0136313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong FP, 2018. Tau proteins and tauopathies in Alzheimer’s disease. Cell Mol. Neurobiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, 2016. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc. Natl. Acad. Sci. U S A 113, 4753–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claerhout S, Dutta B, Bossuyt W, Zhang F, Nguyen-Charles C, Dennison JB, Yu Q, Yu S, Balázsi G, Lu Y, Mills GB, 2012. Abortive autophagy induces endoplasmic reticulum stress and cell death in cancer cells. PLoS One 7, e39400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm PM, Spiegel J, Grossmann TN, Waldmann H, 2015. Direct modulation of small GTPase activity and function. Angew. Chem. Int. Ed. Engl 54, 13516–13537. [DOI] [PubMed] [Google Scholar]
- Cureton DK, Burdeinick-Kerr R, Whelan SP, 2012. Genetic inactivation of COPI coatomer separately inhibits vesicular stomatitis virus entry and gene expression. J. Virol 86, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custer SK, Todd AG, Singh NN, Androphy EJ, 2013. Dilysine motifs in exon 2b of SMN protein mediate binding to the COPI vesicle protein alphα-COP and neurite outgrowth in a cell culture model of spinal muscular atrophy. Hum. Mol. Genet 22, 4043–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custer SK, Foster JN, Astroski JW, Androphy EJ, 2019. Abnormal Golgi morphology and decreased COPI function in cells with low levels of SMN. Brain Res. 1706, 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daro E, Sheff D, Gomez M, Kreis T, Mellman I, 1997. Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component epsilon-COP. J. Cell Biol 139, 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YQ, Xu GG, Duan P, Zhang Q, Wang JZ, 2005. Effects of melatonin on wortmannin-induced tau hyperphosphorylation. Acta Pharmacol. Sin 26, 519–526. [DOI] [PubMed] [Google Scholar]
- Esler WP, Wolfe MS, 2001. A portrait of Alzheimer secretases–new features and familiar faces. Science 293, 1449–1454. [DOI] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R, 2000. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 19, 3905–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI, 2009. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem 284, 12845–12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman JL, Holmes BB, Diamond MI, 2015. Sensitive detection of proteopathic seeding activity with FRET flow cytometry. J. Vis. Exp e53205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, Kama R, Gerst JE, 2007. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol. Cell Biol 27, 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, 1991. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349, 704–706. [DOI] [PubMed] [Google Scholar]
- Guo Q, Penman M, Trigatti BL, Krieger M, 1996. A single point mutation in epsilon-COP results in temperature-sensitive, lethal defects in membrane transport in a Chinese hamster ovary cell mutant. J. Biol. Chem 271, 11191–11196. [DOI] [PubMed] [Google Scholar]
- Guttula SV, Allam A, Gumpeny RS, 2012. Analyzing microarray data of Alzheimer’s using cluster analysis to identify the biomarker genes. Int. J. Alzheimers Dis 2012, 649456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KA, Oyler GA, Doolittle GM, Vincent I, Lehman RA, Kincaid RL, Billingsley ML, 1993. Okadaic acid induces hyperphosphorylated forms of tau protein in human brain slices. Ann. Neurol 33, 77–87. [DOI] [PubMed] [Google Scholar]
- Ho YS, Yang X, Lau JC, Hung CH, Wuwongse S, Zhang Q, Wang J, Baum L, So KF, Chang RC, 2012. Endoplasmic reticulum stress induces tau pathology and forms a vicious cycle: implication in Alzheimer’s disease pathogenesis. J. Alzheimers Dis 28, 839–854. [DOI] [PubMed] [Google Scholar]
- Huang GN, 2012. Biotinylation of cell surface proteins. Bio Protoc. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K, Brett M, Nishi E, Drunat S, Tan ES, Fujiki K, Lebon S, Cham B, Masuda K, Arakawa M, Jacquinet A, Yamazumi Y, Chen ST, Verloes A, Okada Y, Katou Y, Nakamura T, Akiyama T, Gressens P, Foo R, Passemard S, Tan EC, El Ghouzzi V, Shirahige K, 2016. ARCN1 mutations cause a recognizable craniofacial syndrome due to COPI-mediated transport defects. Am. J. Hum. Genet 99, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensson BO, Hansdottir S, Arnadottir GA, Sulem G, Kristjansson RP, Oddsson A, Benonisdottir S, Jonsson H, Helgason A, Saemundsdottir J, Magnusson OT, Masson G, Thorisson GA, Jonasdottir A, Jonasdottir A, Sigurdsson A, Jonsdottir I, Petursdottir V, Kristinsson JR, Gudbjartsson DF, Thorsteinsdottir U, Arngrimsson R, Sulem P, Gudmundsson G, Stefansson K, 2017. COPA syndrome in an Icelandic family caused by a recurrent missense mutation in COPA. BMC Med. Genet 18, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Wang L, Guan Y, Xu H, Niu Y, Han L, Wei YP, Lin L, Chu J, Wang Q, Yang Y, Pei L, Wang JZ, Tian Q, 2014. Golgin-84-associated Golgi fragmentation triggers tau hyperphosphorylation by activation of cyclin-dependent kinase-5 and extracellular signal-regulated kinase. Neurobiol. Aging 35, 1352–1363. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, IbrahimVerbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K, 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med 368, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstrom H, Bergman A, Lendahl U, Näslund J, Lundkvist J, 2002. A sensitive and quantitative assay for measuring cleavage of presenilin substrates. J. Biol. Chem 277, 6763–6766. [DOI] [PubMed] [Google Scholar]
- Knopman DS, Jack CR, Wiste HJ, Weigand SD, Vemuri P, Lowe VJ, Kantarci K, Gunter JL, Senjem ML, Mielke MM, Roberts RO, Boeve BF, Petersen RC, 2013. Brain injury biomarkers are not dependent on betaamyloid in normal elderly. Ann. Neurol 73, 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Mou X, Zhi X, Zhang X, Yang Y, 2014. Dynamic regulatory network reconstruction for Alzheimer’s disease based on matrix decomposition techniques. Comput. Math. Methods Med 2014, 891761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ber I, De Septenville A, Guerreiro R, Bras J, Camuzat A, Caroppo P, Lattante S, Couarch P, Kabashi E, Bouya-Ahmed K, Dubois B, Brice A, 2014. Homozygous TREM2 mutation in a family with atypical frontotemporal dementia. Neurobiol. Aging 35, 2419 e23–2419 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Custer SK, Gilson T, Hao le T., Beattie CE, Androphy EJ, 2015. alphα-COP binding to the survival motor neuron protein SMN is required for neuronal process outgrowth. Hum. Mol. Genet 24, 7295–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZC, Fu ZQ, Song J, Zhang JY, Wei YP, Chu J, Han L, Qu N, Wang JZ, Tian Q, 2012. Bip enhanced the association of GSK-3beta with tau during ER stress both in vivo and in vitro. J. Alzheimers Dis 29, 727–740. [DOI] [PubMed] [Google Scholar]
- Ma Y, 2020. Temporal quantitative profiling of newly synthesized proteins during Abeta accumulation. J. Proteome Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NV, Desjardins A, Leclerc N, 2017. Tau secretion is correlated to an increase of Golgi dynamics. PLoS One 12, e0178288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L, 2011. The many faces of tau. Neuron 70, 410–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multhaup G, Huber O, Buée L, Galas MC, 2015. Amyloid precursor protein (APP) metabolites APP intracellular fragment (AICD), Abeta42, and tau in nuclear roles. J. Biol. Chem 290, 23515–23522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen NP, Kysenius K, Sakha P, Tammela P, Huttunen HJ, 2012. Gamma-Aminobutyric acid type A (GABAA) receptor activation modulates tau phosphorylation. J. Biol. Chem 287, 6743–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski CM, Urano F, 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490, 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjaerg L, Konttinen Y, Peltonen L, 2002. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet 71, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Spencer CH, 2019. An unprecedented COPA gene mutation in two patients in the same family: comparative clinical analysis of newly reported patients with other known COPA gene mutations. Pediatr. Rheumatol. Online J 17, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter CJ, Evans M, Thayanithy V, Taniguchi-Ishigaki N, Bach I, Kolpak A, Bassell GJ, Rossoll W, Lorson CL, Bao ZZ, Androphy EJ, 2011. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum. Mol. Genet 20, 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, 2019. Why Abeta42 is much more toxic than Abeta40. ACS Chem. Neurosci 10, 2843–2847. [DOI] [PubMed] [Google Scholar]
- Piras A, Collin L, Grüninger F, Graff C, Rönnbäck A, 2016. Autophagic and lysosomal defects in human tauopathies: analysis of post-mortem brain from patients with familial Alzheimer disease, corticobasal degeneration and progressive supranuclear palsy. Acta Neuropathol. Commun 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Michnick SW, 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979. [DOI] [PubMed] [Google Scholar]
- Sackmann C, Hallbeck M, 2020. Oligomeric amyloid-beta induces early and widespread changes to the proteome in human iPSC-derived neurons. Sci. Rep 10, 6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Akiyama M, Araki Y, Sumioka A, Shiono M, Taru H, Nakaya T, Yamamoto T, Suzuki T, 2011. Intracellular trafficking of the amyloid beta-protein precursor (APP) regulated by novel function of X11-like. PLoS One 6, e22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, Miller TM, Grinberg LT, Seeley WW, Diamond MI, 2014. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selivanova A, Winblad B, Farmery MR, Dantuma NP, Ankarcrona M, 2006. COPI-mediated retrograde transport is required for efficient gamma-secretase cleavage of the amyloid precursor protein. Biochem. Biophys. Res. Commun 350, 220–226. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ, Hardy J, 2016. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med 8, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis DW, Aparicio RE, Schekman R, 2017. Neurodegeneration-associated mutant TREM2 proteins abortively cycle between the ER and ER-Golgi intermediate compartment. Mol. Biol. Cell 28, 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopschinski BE, Thomas TL, Nadji S, Darvish E, Fan L, Holmes BB, Modi AR, Finnell JG, Kashmer OM, Estill-Terpack S, Mirbaha H, Luu HS, Diamond MI, 2020. A synthetic heparinoid blocks Tau aggregate cell uptake and amplification. J. Biol. Chem 295, 2974–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Xie N, Tang B, Li R, Shen Y, 2017. Alzheimer’s disease: from genetic variants to the distinct pathological mechanisms. Front Mol. Neurosci 10, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo AG, Bharti A, Trujillo C, Harrison R, Murphy JR, 2008. COPI coatomer complex proteins facilitate the translocation of anthrax lethal factor across vesicular membranes in vitro. Proc. Natl. Acad. Sci. U S A 105, 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumuraya T, Matsushita M, 2014. COPA and SLC4A4 are required for cellular entry of arginine-rich peptides. PLoS One 9, e86639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich JD, Ulland TK, Colonna M, Holtzman DM, 2017. Elucidating the role of TREM2 in Alzheimer’s disease. Neuron 94, 237–248. [DOI] [PubMed] [Google Scholar]
- Vivithanaporn P, Yan S, Swanson GT, 2006. Intracellular trafficking of KA2 kainate receptors mediated by interactions with coatomer protein complex I (COPI) and 14–3-3 chaperone systems. J. Biol. Chem 281, 15475–15484. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhai Y, Pang X, Niu T, Ding YH, Dong MQ, Hsu VW, Sun Z, Sun F, 2016. Structural characterization of coatomer in its cytosolic state. Protein Cell 7, 586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Balaji V, Kaniyappan S, Krüger L, Irsen S, Tepper K, Chandupatla R, Maetzler W, Schneider A, Mandelkow E, Mandelkow EM, 2017. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener 12, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkin LB, Jessen B, Wiszniewski W, Vece TJ, Jan M, Sha Y, Thamsen M, Santos-Cortez RL, Lee K, Gambin T, Forbes LR, Law CS, Stray-Pedersen A, Cheng MH, Mace EM, Anderson MS, Liu D, Tang LF, Nicholas SK, Nahmod K, Makedonas G, Canter DL, Kwok PY, Hicks J, Jones KD, Penney S, Jhangiani SN, Rosenblum MD, Dell SD, Waterfield MR, Papa FR, Muzny DM, Zaitlen N, Leal SM, Gonzaga-Jauregui C, au, fnm, Boerwinkle E, Eissa NT, Gibbs RA, Lupski JR, Orange JS, Shum AK, 2015. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat. Genet 47, 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann S, Nicholls S, Takeda S, Fan Z, Hyman BT, 2016. Formation, release, and internalization of stable tau oligomers in cells. J. Neurochem 139, 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Kedlaya R, Higuchi H, Ikeda S, Justice MJ, Setaluri V, Ikeda A, 2017. COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kedlaya R, Higuchi H, Ikeda S, Justice MJ, Setaluri V, Ikeda A, 2010. Mutation in archain 1, a subunit of COPI coatomer complex, causes diluted coat color and Purkinje cell degeneration. Plos Genet. 6, e1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Nykänen NP, Brunello CA, Haapasalo A, Hiltunen M, Uronen RL, Huttunen HJ, 2016. FRMD4A-cytohesin signaling modulates the cellular release of tau. J. Cell Sci 129, 2003–2015. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang X, Ju W, Sun L, Zhang H, 2019. Genetic and expression analysis of COPI genes and Alzheimer’s disease susceptibility. Front Genet. 10, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Khandelwal PJ, Chakraborty R, Cuellar TL, Sarangi S, Patel SA, Cosentino CP, O’Connor M, Lee JC, Tanzi RE, Saunders AJ, 2007. An AICD-based functional screen to identify APP metabolism regulators. Mol. Neurodegener 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]