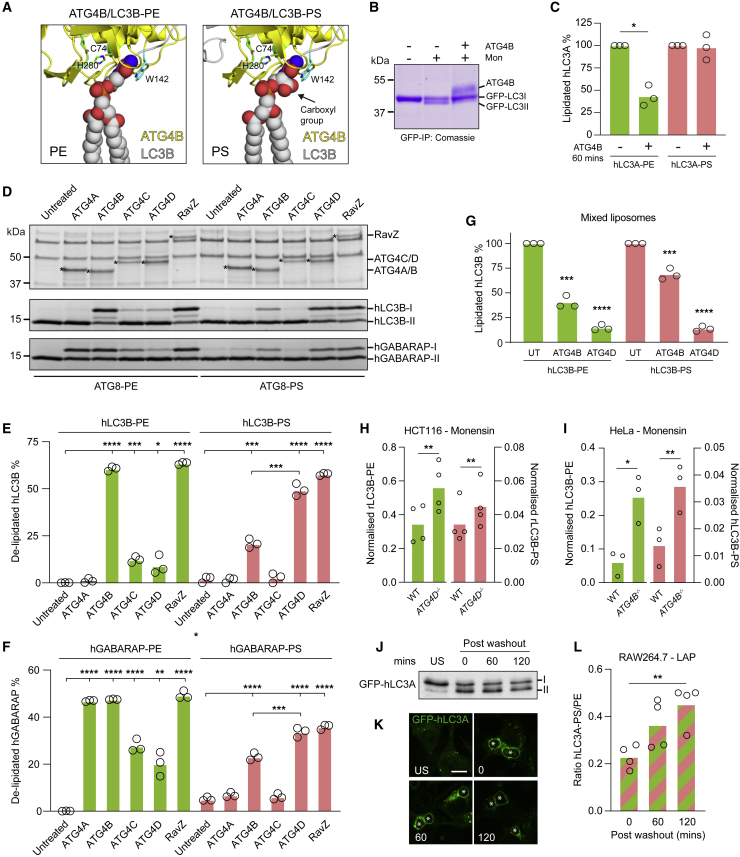
Figure 4.
ATG8-PS and ATG8-PE undergo differential delipidation by the ATG4 family
(A) Molecular modeling of LC3B-PE and LC3B-PS in complex with ATG4B (on the basis of PDB: 2Z0D), with critical catalytic residues marked.
(B) Coomassie staining of GFP-hLC3A IPs from MCF10A ATG13−/− cells −/+ monensin, incubated −/+ ATG4B for 60 min.
(C) Mass spectrometry analysis of hLC3A-PE and hLC3A-PS from cells treated as in (B). Data represent three independent experiments with means normalized to time 0. ∗p < 0.01, paired t test.
(D) PE or PS liposome-based delipidation assays with purified ATG4s or RavZ. Conjugated hLC3B or hGABARAP was incubated with ATG4A/B/C/D/RavZ (asterisk) for 60 min and analyzed using SDS-PAGE/Coomassie.
(E and F) Densitometry analysis of (D). Data represent means from three independent experiments ∗p < 0.03, ∗∗p < 0.002, ∗∗∗p < 0.0002, and ∗∗∗∗p < 0.0001, unpaired t test.
(G) Mass spectrometry analysis of hLC3B conjugation on mixed liposomes, incubated with ATG4B or ATG4D for 60 min. Data represent means normalized to untreated controls from three independent experiments. ∗∗∗p < 0.0002 and ∗∗∗∗p < 0.0001, unpaired t test.
(H and I) Normalized mass spectrometry analysis of GFP-rLC3B from monensin treated WT and ATG4D−/− HCT116 cells (H) and of GFP-hLC3BG120 from monensin treated WT and ATG4B−/− HeLa cells (I). Data represent means from three or four independent experiments. ∗p < 0.03 and ∗∗p < 0.002, paired t test.
(J) Western blot analysis of RAW264.7 cells expressing GFP-hLC3A treated −/+ zymosan for 25 min, followed by washout 0–120 min post-LAP.
(K) Confocal images of cells treated as in (J). Scale bar: 5 μm. Asterisks denote phagosomes.
(L) Ratios of hLC3A-PS/PE measured by mass spectrometry from cells treated as in (J). Data represent means from four independent experiments. ∗∗p < 0.002, unpaired t test.
See also Figure S4.