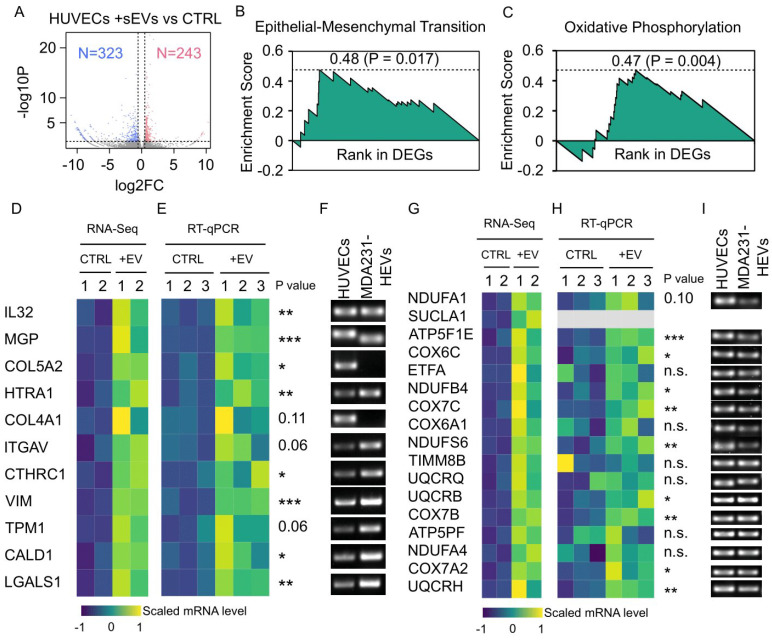
Figure 6.
RNA-seq analysis of HUVECs treated with cancer EVs. (A) Volcano plot shows RNA-seq data of HUVECs with or without the treatment by 50 µg/mL MDA231-HEVs. The fold change and P value were analyzed by HISAT2-HTSeq-DESeq2 pipeline. Differentially expressed genes (DEGs) were highlighted in red for 242 upregulated genes and blue for 323 downregulated genes. Gene set enrichment analysis of the DEGs revealed that EMT gene set (B) and OP gene set (C) were significantly enriched, with enrichment scores of 0.475 (P = 0.017) and 0.471 (P = 0.004), respectively. (D) Heatmap shows the RNA-seq data of upregulated DEGs that were annotated by EMT gene set. (E) RT-qPCR verified the upregulation of the DEGs in (D). (F) DNA gel electrophoresis demonstrated that, except for COL5A2 and COL4A1, the mRNAs of all the DEGs in (D) (9/11) were present in MDA231-HEVs. (G) Heatmap shows the RNA-seq data of upregulated DEGs that were annotated by OP gene set. (H) RT-qPCR verified the upregulation of the DEGs in (G). (I) DNA gel electrophoresis demonstrated that, except for SUCLA1, the mRNAs of all the DEGs in (G) (15/16) were present in MDA231-HEVs. n.s.: not significant, * P < 0.05, ** P < 0.01, *** P < 0.001, based on Student’s t test for (E,H).