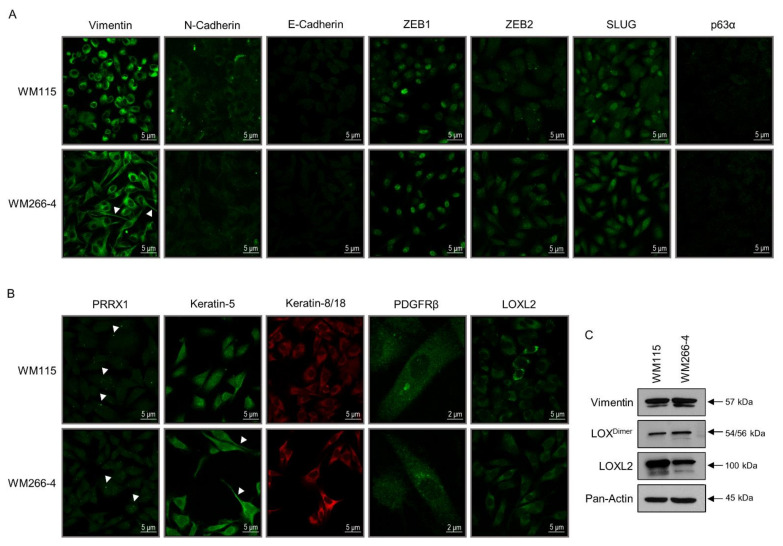
Figure 2.
Engagement of hybrid EMT/MET programs during BRAFV600D-dependent melanomagenesis. (A,B) Immunofluorescence profiles of epithelial (MET) and mesenchymal (EMT) program protein markers in WM115 (primary) and WM266-4 (metastatic) melanoma cells. (C) Western blotting-derived patterns of protein expression being critically implicated in the acquisition of epithelial or mesenchymal (molecular/phenotypic) traits. (A) “Vimentin” (M), “N-Cadherin” (M), “E-Cadherin” (E), “ZEB1” (M), “ZEB2” (M), “SLUG” (M) and “p63α” (E). (B) “PRRX1” (M), “Keratin-5” (E), “Keratin-8/18” (E/M), “PDGFRβ” (E/M), and “LOXL2” (M). (C) “Vimentin” (M), “LOX” (M), “LOXL2” (M), and “Pan-Actin” (protein of reference). E: Epithelial (MET) marker. M: Mesenchymal (EMT) marker. Arrowheads: (A) “Vimentin”-positive Invadopodia and (B) “PRRX1”-positive globular “specks”; “Keratin-5”-positive Invadopodia. (A,B) Scale bars: 2 or 5 μm. (C) Molecular weights of identified proteins (major bands) are denoted by numbers at the right side of each respective panel. Protein quantification values (in bar-chart format) are shown in Figure S1.