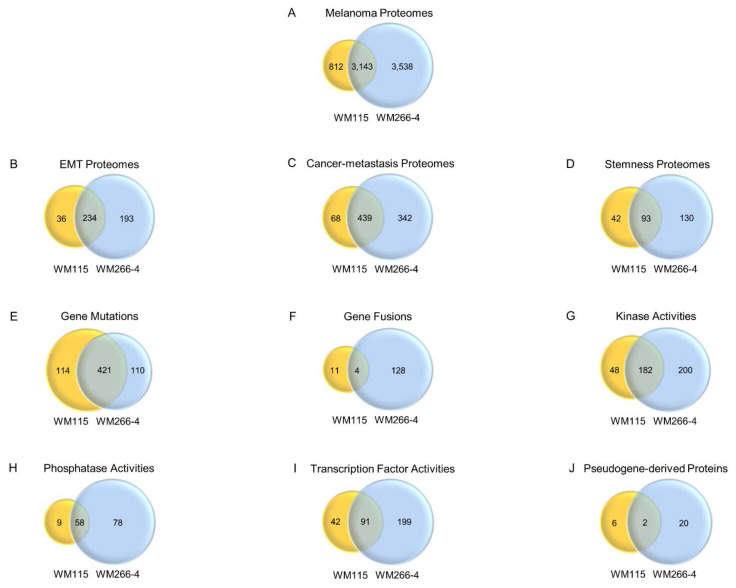
Figure 10.
Proteomic numeration of human BRAFV600D-positive melanoma cells undergoing metastasis. (A–J) Venn diagrams describing the unique and common (overlapped circle areas) proteomic catalogues (protein collections) of WM115 (primary) (yellow-colored) and WM266-4 (metastatic) (blue-colored) BRAFV600D-dependent melanoma cells, according to their (A) total “Melanoma Proteomes” (nLC-MS/MS; UNIPROT [50]; this study and [35]), (B) “EMT Proteomes” (dbEMT; [58,59]), (C) “Cancer-metastasis Proteomes” (CMGene; [60]), (D) “Stemness Proteomes” (LifeMap Discovery; [61]), (E) “Gene Mutations” (DepMapPortal; [62,63,64,65]), (F) “Gene Fusions” (DepMapPortal; [62,63,64,65]), (G) “Kinase Activities” (GO_MF DAVID 6.8; [51,52,53]), (H) “Phosphatase Activities” (GO_MF DAVID 6.8; [51,52,53]), (I) “Transcription Factor Activities” (GO_MF DAVID 6.8; [51,52,53]), and (J) “Pseudogene-derived Proteins” (nLC-MS/MS; UNIPROT [50]; this study and [35]). (D) Venn diagram of shared genes expressed or selectively not expressed in human embryonic stem cells (hESCs) between WM115 (primary) and WM266-4 (metastatic) human melanoma cells. The genes involved in this analysis were obtained from LifeMap Discovery [61] and were manually curated from the literature and/or imported from high-throughput experiments. (F) Note the remarkably increased number (~11.5×) of unique “Gene Fusions” in WM266-4 (metastatic) (n = 128) (Table S19) compared to the respective WM115 (primary) (n = 11) (Table S18) melanoma cells ones.