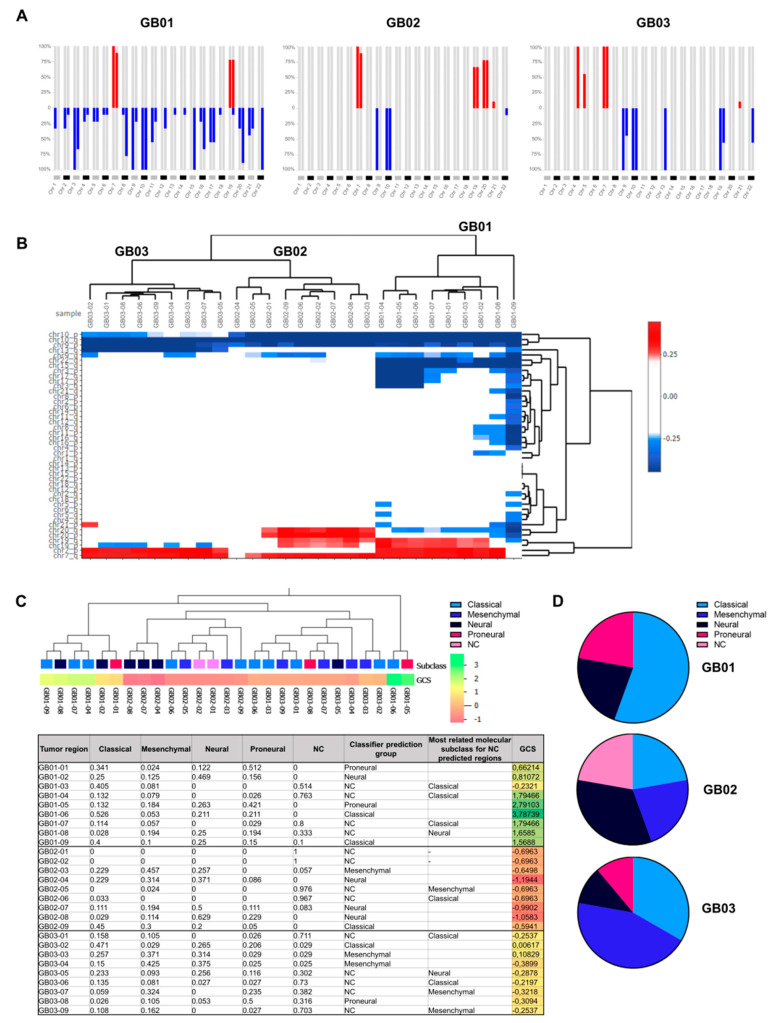
Figure 2.
Copy number variation (CNV) analysis and molecular subtype correlation. (A) CNApp frequencies for the p and q arms of each chromosome. Alteration frequency is expressed as the percentage of altered regions out of the total of 9 regions within each tumor (red for gains and blue for losses). (B) Hierarchical clustering of copy number variations of chromosomal regions (p and q arms). Hierarchical cluster analysis according to Pearson’s correlation of the three GB samples subdivided into their intra-tumoral regions. (C) Correlation with the four molecular subtypes and hierarchical clustering using the random forests algorithm. The CNApp classifier model was applied to our three GBs and 480 GBs derived from the TCGA-GBM data collection with molecular subclass annotation. Tumor regions in our 3 GBs were included in the classifier as “not classified” (NC) and correlation was performed using the global score (GCS) that CNApp assigns during resegmentation, by which it classifies and weights CNVs based on their length and width. The values in the table are the correlation coefficients that each region has with each of the four molecular subclasses and with the NC class. The “Classifier prediction group” column reports the molecular subtype that the system has correlated and predicted. The “Most related molecular subclass for NC predicted regions” column reports the best correlation found with one of the four molecular subclasses when the tumor region was associated with NC by the classifier. (D) Pie chart of the molecular composition of each tumor. Each tumor is composed of several molecular subclasses, each associated with an intra-tumoral region.