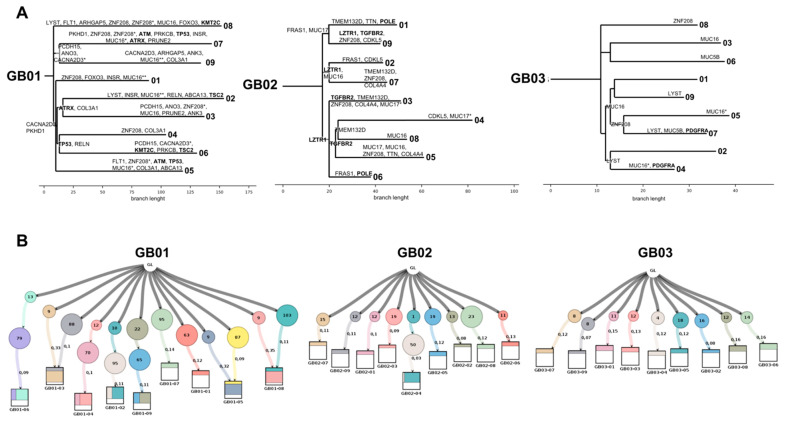
Figure 6.
Tumor phylogeny and clonal evolution. (A) Phylogenetic trees. For each sample, a rooted tree was created whose leaf nodes are tumor regions. The length of the branches is equal to the number of mutations. Genes annotated on the tree are those with which the system has distinguished (through the absence or presence of specific mutations) and divided the different leaf branches representing the 9 tumor portions. Genes in bold are driver genes. (B) Clonal evolution lineage tree and sample composition. The lineage tree was built based on the constraint network using Lineage Inference for Cancer Heterogeneity and Evolution (LICHeE). Each node (circle) represents a subpopulation of GB cells. All nodes arose from a single hypothetical clone called Germline (GL), representing the genetic architecture of normal tissue from the same patient. Numbers within the circles indicate the number of nucleotide variants shared by the cluster; numbers outside the circles show the average variant allele frequency (VAF) of the variants in each cluster. Squares represent each individual tumor region, with colored rectangles indicating the cell fraction represented by the clonal cluster.