Table 1.
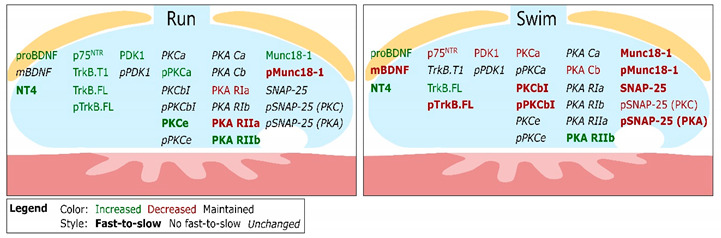
Summary of exercise-induced molecular adaptations. (A) Fast-to-slow transition following either running or swimming training protocols. In the table, blue-shaded cells highlight the values that after the training protocol are statistically similar to those in sedentary mice soleus muscles. Yellow highlights the values that after the training protocol are opposite to those in sedentary mice soleus, being statistically different from both untrained soleus and plantaris. Notes: Lighter colors correspond to these changes induced by trainings despite that no significative differences were found between untrained plantaris and soleus muscles. To facilitate data analysis, the first column corresponds to the values of soleus muscles in relation with plantaris (both untrained) that were published for first time in [32]. Statistical significance of the differences between the experimental groups was evaluated under a non-parametric Kruskal–Wallis test followed by Dunn’s post hoc test. The criterion for statistical significance against WT plantaris was as follows: * p < 0.05, ** p < 0.01, and *** p < 0.001 (precise p. value is provided). (B) Summary of the results of this work. Representation of the molecular changes in the BDNF/TrkB signaling pathways after run (left) and swim (right) training protocols. The figure shows that molecular changes are more abundant and mainly in the fast-to-slow direction after the swimming protocol, due to the intensity-dependent effect.
| A | SOL WT | PLA WT Run | PLA WT Swim | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NTFs and receptors | proBDNF | 1.05 | ± | 0.31 | 0.921 | 2.65 | ± | 0.89 | 0.008 | ** | 2.17 | ± | 0.93 | 0.045 | * | |
| mBDNF | 0.57 | ± | 0.27 | 0.021 | * | 0.95 | ± | 0.29 | 0.736 | 0.45 | ± | 0.18 | 0.002 | ** | ||
| NT4 | 1.89 | ± | 0.03 | 0.000 | *** | 1.81 | ± | 0.10 | 0.000 | *** | 1.51 | ± | 0.14 | 0.000 | *** | |
| p75NTR | 1.11 | ± | 0.03 | 0.393 | 1.68 | ± | 0.24 | 0.001 | ** | 0.79 | ± | 0.10 | 0.006 | ** | ||
| TrkB.T1 | 0.92 | ± | 0.06 | 0.619 | 2.45 | ± | 0.14 | 0.000 | *** | 0.92 | ± | 0.14 | 0.305 | |||
| TrkB.FL | 0.63 | ± | 0.04 | 0.012 | ** | 1.95 | ± | 0.62 | 0.022 | * | 1.84 | ± | 0.54 | 0.021 | * | |
| pTrkB.FL (Y816) | 0.19 | ± | 0.01 | 0.000 | *** | 1.23 | ± | 0.14 | 0.014 | * | 0.40 | ± | 0.16 | 0.000 | *** | |
| Serine-threonine kinases | PDK1 | 1.35 | ± | 0.09 | 0.089 | 1.94 | ± | 0.51 | 0.033 | * | 0.62 | ± | 0.09 | 0.002 | ** | |
| pPDK1 (S241) | 0.99 | ± | 0.26 | 0.998 | 0.96 | ± | 0.08 | 0.452 | 0.99 | ± | 0.05 | 0.824 | ||||
| cPKCα | 1.19 | ± | 0.01 | 0.258 | 0.81 | ± | 0.22 | 0.222 | 0.68 | ± | 0.15 | 0.020 | * | |||
| pcPKCα (S657) | 0.61 | ± | 0.03 | 0.014 | ** | 1.42 | ± | 0.18 | 0.008 | ** | 0.95 | ± | 0.20 | 0.688 | ||
| cPKCβI | 0.15 | ± | 0.11 | 0.000 | *** | 0.99 | ± | 0.53 | 0.976 | 0.53 | ± | 0.23 | 0.023 | * | ||
| pcPKCβI (T621) | 0.51 | ± | 0.22 | 0.000 | *** | 1.11 | ± | 0.18 | 0.328 | 0.56 | ± | 0.08 | 0.000 | *** | ||
| nPKCε | 1.65 | ± | 0.32 | 0.000 | *** | 1.41 | ± | 0.28 | 0.017 | * | 0.76 | ± | 0.20 | 0.099 | ||
| pnPKCε (S729) | 0.77 | ± | 0.06 | 0.140 | 0.89 | ± | 0.26 | 0.508 | 0.73 | ± | 0.20 | 0.085 | ||||
| PKA Cα | 2.04 | ± | 0.07 | 0.000 | *** | 1.09 | ± | 0.14 | 0.270 | 1.29 | ± | 0.42 | 0.215 | |||
| PKA Cβ | 1.13 | ± | 0.43 | 0.648 | 1.09 | ± | 0.11 | 0.147 | 0.60 | ± | 0.24 | 0.014 | * | |||
| PKA RIα | 2.4 | ± | 0.56 | 0.000 | *** | 0.77 | ± | 0.16 | 0.032 | * | 1.18 | ± | 0.37 | 0.364 | ||
| PKA RIβ | 1.15 | ± | 0.01 | 0.776 | 0.98 | ± | 0.22 | 0.846 | 0.83 | ± | 0.31 | 0.312 | ||||
| PKA RIIα | 0.79 | ± | 0.09 | 0.583 | 0.76 | ± | 0.16 | 0.027 | * | 0.94 | ± | 0.19 | 0.553 | |||
| PKA RIIβ | 1.98 | ± | 0.07 | 0.006 | ** | 1.76 | ± | 0.41 | 0.013 | * | 1.34 | ± | 0.28 | 0.049 | * | |
| Exocytosis machinery | Munc18-1 | 0.75 | ± | 0.12 | 0.081 | 1.30 | ± | 0.06 | 0.000 | *** | 0.47 | ± | 0.12 | 0.000 | *** | |
| pMunc18-1 (S313) | 0.54 | ± | 0.11 | 0.000 | *** | 0.65 | ± | 0.17 | 0.004 | ** | 0.70 | ± | 0.18 | 0.017 | * | |
| SNAP-25 | 0.75 | ± | 0.17 | 0.050 | 1.06 | ± | 0.17 | 0.515 | 0.40 | ± | 0.08 | 0.000 | *** | |||
| pSNAP-25 (S187) | 2.31 | ± | 0.06 | 0.000 | *** | 1.03 | ± | 0.15 | 0.700 | 0.47 | ± | 0.07 | 0.000 | *** | ||
| pSNAP-25 (T138) | 0.35 | ± | 0.21 | 0.000 | *** | 1.27 | ± | 0.19 | 0.158 | 0.53 | ± | 0.16 | 0.002 | ** | ||
| B |
|
|||||||||||||||
