Figure 7.
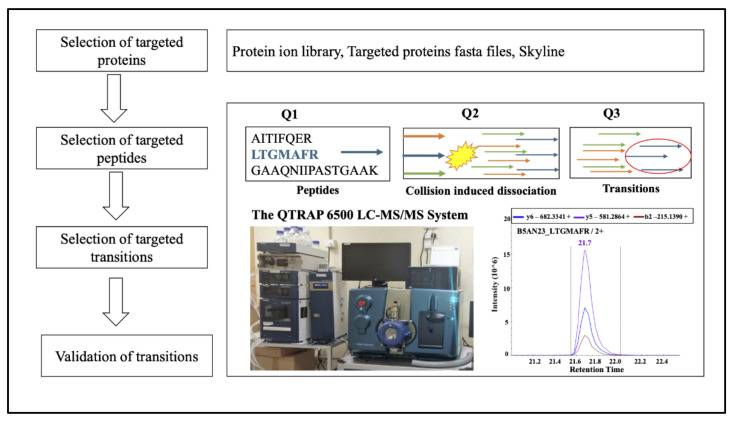
Schematic workflow of quantitative MRM-based targeted proteomics in myopic vs. control eyes (n = 5). To establish the MRM acquisition method, proteins of interest were selected based on the untargeted SWATH datasets. The targeted peptides were firstly checked and extracted from the previously acquired MS spectral IDA library identified by the ProteinPilot. The whole sequence of targeted proteins in FASTA format was first downloaded from the UniProt and then imported to the Skyline software (MacCoss Lab, Seattle, WA, USA). The top one transition of GAPDH was selected with the largest peak area and considered as an internal standard transition. The digested peptides (2µg) from each retinal sample were injected into QTRAP 6500. The MRM raw data were further processed MultiQuantTM. The MQ4 algorithm for automatic peak integration.