Figure 2.
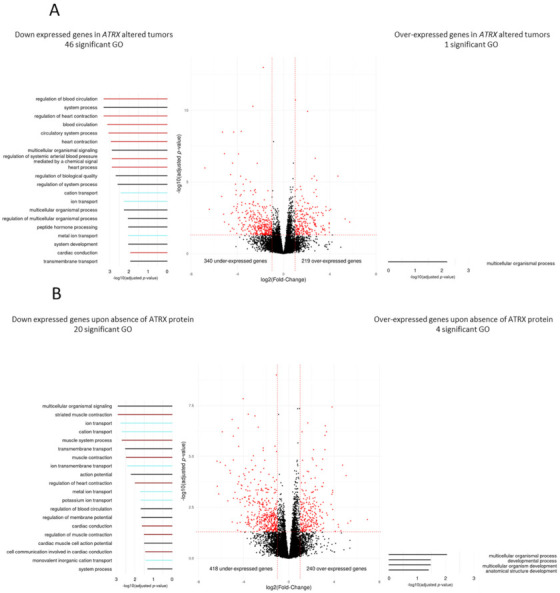
Differential gene expression and Gene Ontology analyses according to ATRX alteration in leiomyosarcomas (Cohort 1). Differentially expressed genes according to (A) ATRX status (wild-type vs. altered) or (B) ATRX expression (nucleus vs. absent). Red dots indicate significant genes (p ≤ 0.05 and fold-change ≤−2 or ≥2). Gene Ontology (GO) analyses, represented on the left (under-expressed genes) and the right (over-expressed genes), identified 46 and 1 significant GO terms (p ≤ 0.05), respectively in (A) and 20 and 4 significant GO terms (p ≤ 0.05) in (B). On each side, the 20 most significant GO terms are represented and color-coded by mechanisms; light red, dark red, light blue, and black colors indicate “circulatory system process”, “muscle system process”, “ion transport” and general terms, respectively. For ATRX localization, the “absent” group means “not at the nucleus”, including all cases without expression and the case with a cytoplasmic localization (LMS16). All p-values were adjusted by Benjamini and Hochberg method.
