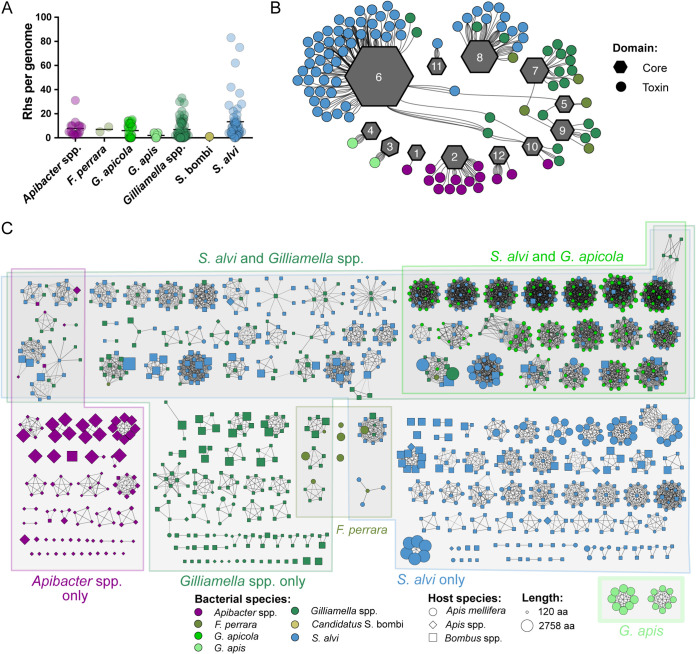
FIG 3.
Rhs toxin diversity within bee gut symbionts. (A) Number of Rhs toxin domains per genome for each species cluster. C-terminal toxin domains were extracted from proteins containing Rhs repeat motifs and clustered. Final counts represent the number of proteins with ≥40% homology to representative toxin domains. (B) Network diagram showing relationships between Rhs toxin domains and core domains. Gray hexagons represent core domain clusters, with the size proportional to the number of proteins within the cluster. Circles represent toxin domain clusters, which are colored based on the bacterial species in which they are most often found. Edges connect core and toxin domains from the same protein. Not all toxin domains are shown, as many orphaned toxin domains do not include a large enough core domain to be assigned to a cluster. (C) Network diagram illustrating the composition of 208 toxin domain clusters. Each node represents a protein sequence from an isolate genome. Node color, shape, and size represent the bacterial species, the host from which it was isolated, and the protein length, respectively. Clusters are arranged based on whether they are found in multiple species clusters or restricted to one genus. Edges represent ≥40% amino acid identity over a minimum of 90 amino acids, as determined through all-versus-all protein BLAST.