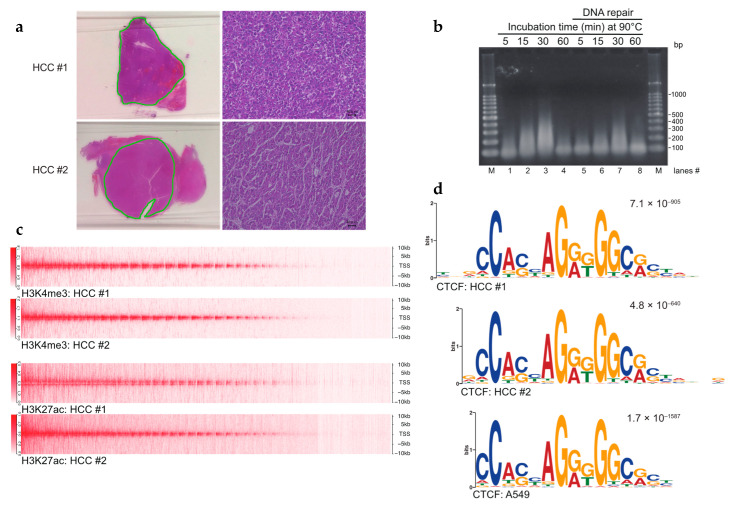
Figure 1.
Establishment of the FFPE ChIP-seq procedure. (a) Hematoxylin and eosin (H&E) staining of hepatocellular carcinoma (HCC) tissues. Dissected tissues utilized for FFPE ChIP-seq are underlined in green. Higher magnification images (scale bars, 40 µm) are shown on the right sides. (b) Controlled heat-treatment of FFPE samples yielded solubilized chromatin. Sectioned FFPE tissues were treated with an overnight incubation at 65 °C followed by indicated time points at 90 °C. Reversed-crosslinked DNAs were repaired in lanes 5–8. Purified DNAs were amplified by whole genome amplification (WGA) and subjected to 1% agarose gel electrophoresis. M: 100-bp DNA ladders. (c) Heat map (upper half) showing H3K4me3 enrichment at TSSs ± 10 kb in HCCs. Heat map (lower half) for H3K27ac enrichment. (d) Sequence logos of CTCF ChIP-seq in HCCs. The top motif identified by MEME-ChIP, algorithms for de novo motif discovery, is shown. E-values, an estimate of the expected number of motifs with the given log-likelihood ratio, are shown on the upper right. Sequence logos in the A549 cell line were obtained from GSE30263.