FIGURE 6.
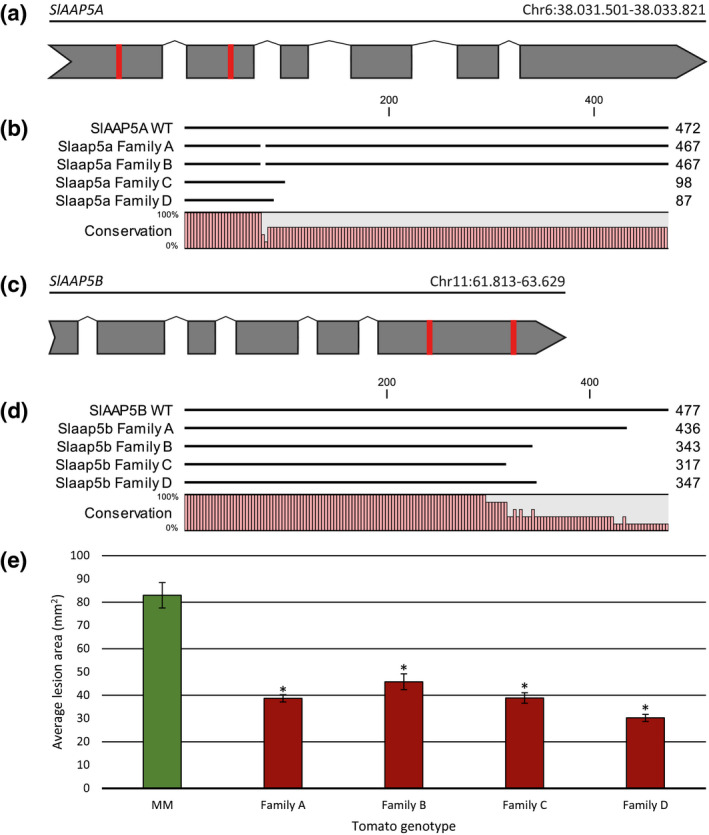
CRISPR‐Cas9 mediated mutagenesis of SlAAP5A and SlAAP5B results in tomato mutants with partial Phytophthora infestans resistance. (a, c) Schematic representations of the gene model of the tomato genes SlAAP5A (a) and SlAAP5B (c). Exons are indicated in grey. 20 bp target sequences for CRISPR‐Cas9 mediated genome editing are indicated in red. (b, d) Multiple sequence alignments of the predicted protein sequences of SlAAP5A (b) and SlAAP5B (d) in four homozygous tomato T2 families with CRISPR‐Cas9‐induced mutations, compared to the reference protein sequence. Data S20 gives the full sequence alignments. (e) Four tomato T2 families with mutations in SlAAP5A and SlAAP5B as indicated in (b) and (d) were tested for susceptibility to P. infestans isolate CA56 compared to wild‐type control cv. Moneymaker (MM). Lesion areas (in mm2) were measured at 4 days postinoculation. Each bar represents the average lesion area of 48–63 biological replicates. Error bars indicate the standard error of the mean. Asterisks indicate significant differences compared to the average lesion area of MM (t test, p < .05)
