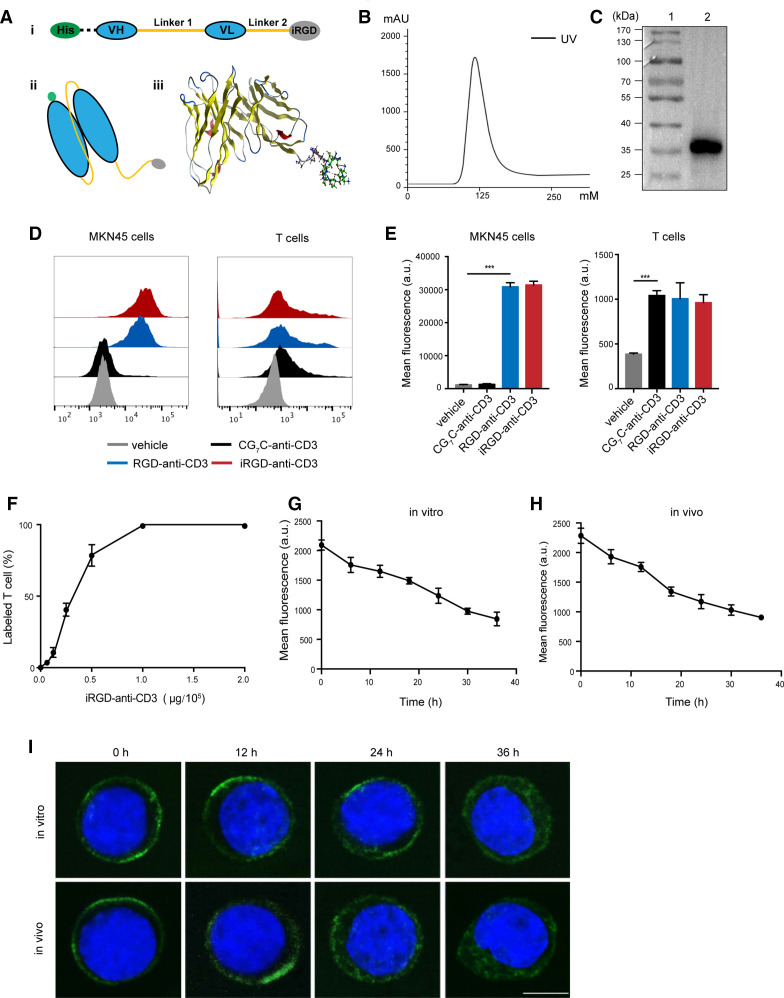
Figure 1.
Generation and binding properties of iRGD-anti-CD3. (A) (i) The design of DNA fragments coding iRGD-anti-CD3. Linker 1, G4S×3; linker 2, G4s; (ii) schematic illustration of iRGD-anti-CD3 structure; (iii) and molecular dynamics simulation using the molecular operating environment 2014 (Montreal, Quebec, Canada). (B) Purification profile of refolded iRGD-anti-CD3. UV, ultraviolet. (C) Eluted fractions were identified by western blot analysis. Lane 1, protein ladder; lane 2, purified iRGD-anti-CD3. (D) Flow cytometry results showing bindings of different reagents to MKN45 and PBMCs. (E) Mean fluorescence intensity on MKN45 and PBMCs in (D). Data are represented as mean±SEM, n=3. Student’s t-test, ***p<0.001. (F) Analysis of the percentage of T cells bound with iRGD-anti-CD3 using flow cytometry. (G) T cells modified with iRGD-anti-CD3 were cultured in vitro for 12–36 hours, and averaged fluorescence intensities of T cells were analyzed by flow cytometry. (H) Mice were injected with T cells modified with iRGD-anti-CD3 via tail vein, and then blood was drawn 12–36 hours after injection. Averaged fluorescence intensities of T cells were analyzed. (I) Representative confocal images of T cells (upper panel, from G; lower panel, from H) at the indicated time points. iRGD-anti-CD3 was detected with Alexa Fluor 488-anti-His tag antibody, green; nucleus, blue. Scale bar, 5 µm. PBMC, peripheral blood mononuclear cell; VH, heavy chain variable domain; VL, light chain variable domain.