Figure 3.
Whole-transcriptome analysis of human ocular surface cells infected by SARS-CoV-2
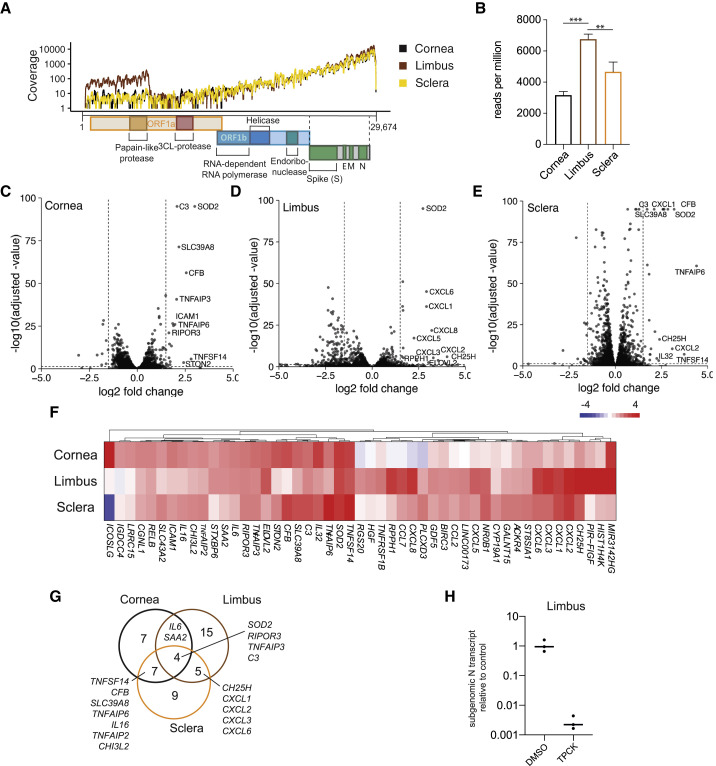
(A) Read coverage of the SARS-CoV-2 genome in human donor cornea, limbus, and sclera tissues infected with SARS-CoV-2 (MOI = 1.0, 24 h) and subjected to mRNA sequencing (mRNA-seq).
(B) Quantification of reads mapped in (A), indicated as mean reads per million. Error bars indicate SD (n = 3/condition); ∗∗adjusted p = 0.0024 and ∗∗∗adjusted p = 0.0001 (one-way ANOVA).
(C–E) Differential gene expression of all genes plotted by the log2 fold change and the statistical significance as −log10 of the adjusted p value the experiment in (A) and (B) in (C) cornea, (D) limbus, and (E) sclera. The ten genes with highest increases in gene expression that also reached statistical significance (adjusted p < 0.05) were named. Horizontal line indicates adjusted p = 0.05, and vertical lines indicate log2 fold change = ±1.5.
(F) Heatmap comparing the differential gene expression of genes that showed increased gene expression upon infection (log2 fold change > 1.5, adjusted p < 0.05) in at least one of the tissues as in (C)–(E).
(G) Venn diagram showing the overlap of genes with increased expression (log2 fold change > 1.5, adjusted p < 0.05) in response to SARS-CoV-2 infection as in (C)–(F).
(H) Relative expression of subgenomic SARS-CoV-2 N gene RNA in infected human donor limbus tissue (MOI = 1.0, 24 h) pre-treated with TPCK or vehicle control (DMSO) quantified by qRT-PCR. Relative expression = 1 is the average expression in the vehicle control (n = 3/condition).
See also Table S1.