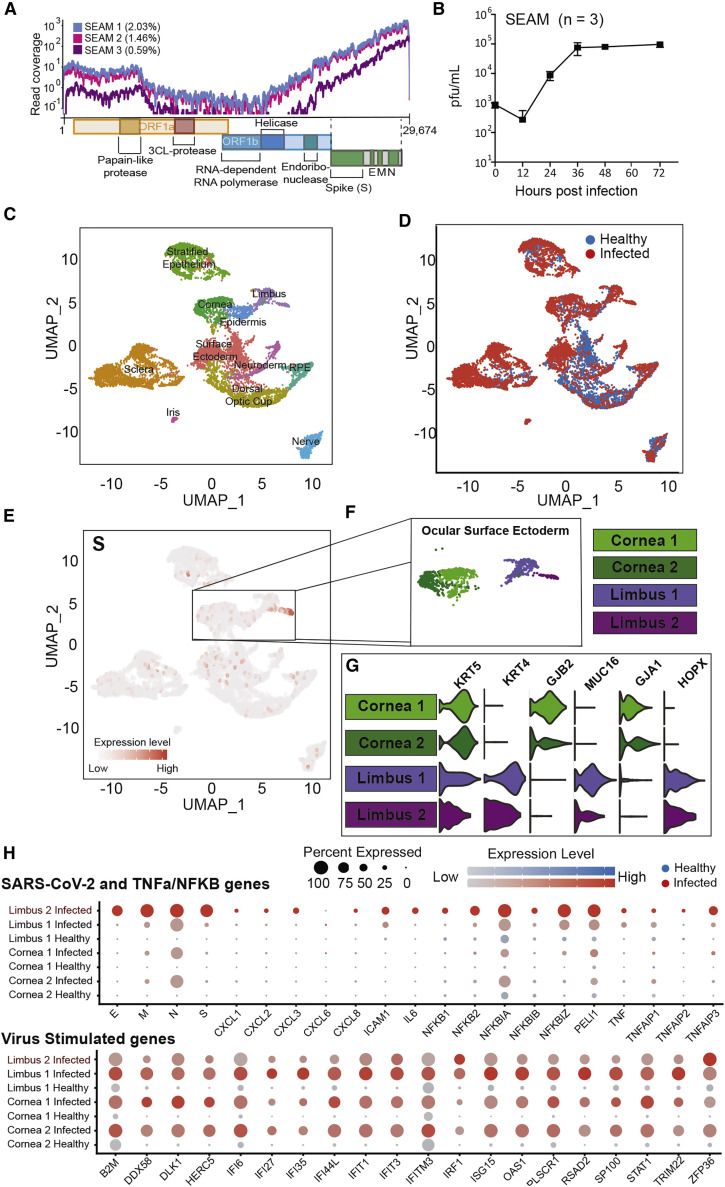
Figure 6.
SARS-CoV-2 infection of SEAM whole-eye colonies
(A) Read coverage of the SARS-CoV-2 genome from next-generation sequencing of RNA from three different infected SEAM cultures (MOI = 1.0, 24 h).
(B) Virus growth in SEAM cultures shown as plaque-forming units per milliliter of supernatant at the time points indicated. Error bars show SD (n = 3).
(C) UMAP based on unbiased clustering of integrated scRNA-seq data from a non-infected SEAM culture (healthy [SEAM differentiated for 31 days]) and a SEAM infected with SARS-CoV-2 (MOI = 1.0, 24 h) (infected [SEAM differentiated for 54 days]), resolution 0.75.
(D) UMAP of integrated scRNA-seq data colored by origin of data (healthy [blue] versus infected [red]).
(E) Feature plot showing the location of cells expressing the SARS-CoV-2 spike gene (S) in red.
(F) Zoom in of the cornea and limbal clusters made up of four sub-clusters.
(G) Violin plots showing some differences in genes expression between the corneal and limbal sub-clusters used for sub-annotation of cell-type identity.
(H) Dot plots showing differential expression of inflammation related genes between the corneal and limbal sub-clusters split by data origin, healthy (blue) versus infected (red). Top: SARS-CoV-2 genes and TNF-α/NF-κB pathway-related gens. Bottom: SARS-CoV-2-stimulated genes.