Significance
Toll/Toll-like receptors are key regulators of the innate immune system in both invertebrates and vertebrates. However, while mammalian Toll-like receptors function as pattern recognition receptors that directly bind microbe-associated ligands, arthropod Tolls are generally thought to be activated by Spätzle cytokines. Here, we show that Toll9 from the silkmoth Bombyx mori functions as a pattern recognition receptor that binds lipopolysaccharide. Our results provide a link for both structural and functional conservation between an insect Toll and a mammalian Toll-like receptor family member.
Keywords: immunity, lipopolysaccharide, ML proteins, MD-2, Toll
Abstract
Toll/Toll-like receptors (TLRs) are key regulators of the innate immune system in both invertebrates and vertebrates. However, while mammalian TLRs directly recognize pathogen-associated molecular patterns, the insect Toll pathway is thought to be primarily activated by binding Spätzle cytokines that are processed from inactive precursors in response to microbial infection. Phylogenetic and structural data generated in this study supported earlier results showing that Toll9 members differ from other insect Tolls by clustering with the mammalian TLR4 group, which recognizes lipopolysaccharide (LPS) through interaction with myeloid differentiation-2 (MD-2)–like proteins. Functional experiments showed that BmToll9 from the silkmoth Bombyx mori also recognized LPS through interaction with two MD-2–like proteins, previously named BmEsr16 and BmPP, that we refer to in this study as BmMD-2A and BmMD-2B, respectively. A chimeric BmToll9–TLR4 receptor consisting of the BmToll9 ectodomain and mouse TLR4 transmembrane and Toll/interleukin-1 (TIR) domains also activated LPS-induced release of inflammatory factors in murine cells but only in the presence of BmMD-2A or BmMD-2B. Overall, our results indicate that BmToll9 is a pattern recognition receptor for LPS that shares conserved features with the mammalian TLR4–MD-2–LPS pathway.
Toll was initially identified in Drosophila melanogaster as an essential gene in dorsal–ventral (DV) axis formation that activates nuclear factor κB (NF-κB) transcription factor signaling (1). Subsequent studies showed that Toll is an integral membrane receptor which has an extracellular domain (ectodomain) with at least 15 leucine-rich repeats (LRRs), a single-pass transmembrane domain, and a cytoplasmic region named the Toll/interleukin-1 (TIR) homology domain because it shares similarities with the mammalian interleukin-1 receptor (IL-1R) (2, 3). Results further indicated that Drosophila Toll (Toll-1) signaling regulates the inducible expression of certain antimicrobial peptide (AMP) genes in response to infection by bacteria or fungi (4, 5).
Since the first human homolog of Drosophila Toll was cloned (6), at least 10 Toll-like receptors (TLRs) have been identified in mammals that play no roles in DV axis formation but regulate a range of innate immune defense functions (7, 8). Mammalian TLRs are further classified as pattern recognition receptors (PRRs), because they bind microbe-associated ligands including lipopolysaccharide (LPS), peptidoglycan (PGN), flagellin, unmethylated CpG DNA, and viral RNAs (9–14). Since identification of Toll-1, eight other Toll genes have been identified in Drosophila, while comparative data indicate that other insects also encode multiple Toll genes (15). Toll-1 is activated during DV axis formation and in response to infection by a cytokine named Spätzle that binds to the ectodomain after processing from an inactive precursor (5). The ectodomains of other insect Toll family members also bind Spätzle ligands (16–18).
Phylogenetic data indicate the TIR domains of most insect Tolls cluster separately from mammalian TIRs (15, 19). The exception is Toll9 members, which differ from other insect Tolls in regard to amino acid sequence and exon/intron organization, and cluster most closely with the mammalian TLR4 clade that binds LPS (13, 15, 19). TLR4 complexes with CD14 and myeloid differentiation-2 (MD-2) on the cell surface (20). TLR4 also binds the MD-2–LPS complex, which results in receptor activation and expression of inflammatory mediators including tumor necrosis factor α (TNF-α), IL-1, and IL-6 (21, 22). In Drosophila, PGN activates Toll-1 by stimulating Spätzle processing, whereas LPS does not (23, 24). In contrast, both ultrapure PGN and LPS activate AMP gene expression in Lepidoptera (moths and butterflies) like Bombyx mori and Manduca sexta (25, 26). The factors involved in LPS signaling are largely unknown although B. mori, M. sexta, and other insects encode MD-2–related lipid-recognition (ML) protein family members (27–29).
We recently identified three ML proteins in B. mori that were originally named BmEsr16, BmPP, and BmML-1, and presented evidence which implicated BmEsr16 in LPS-induced expression of certain AMP genes (27). Given sequence similarities between insect Toll9 members and mammalian TLR4, this study addressed whether Toll9 and ML proteins interact in LPS recognition in B. mori. Our results indicated that BmToll9 recognizes LPS through interactions with BmEsr16 and BmPP, which we rename here BmMD-2A and BmMD-2B, respectively, to indicate their functional and potential evolutionary similarity to mammalian MD-2 proteins. A chimeric BmToll9–TLR4 receptor consisting of the BmToll9 ectodomain and mouse TLR4 transmembrane and TIR domains also activated LPS-induced release of inflammatory factors by bone marrow-derived macrophage (BMDM) cells from TLR4−/− mice but only in the presence of BmMD-2A or BmMD-2B.
Results
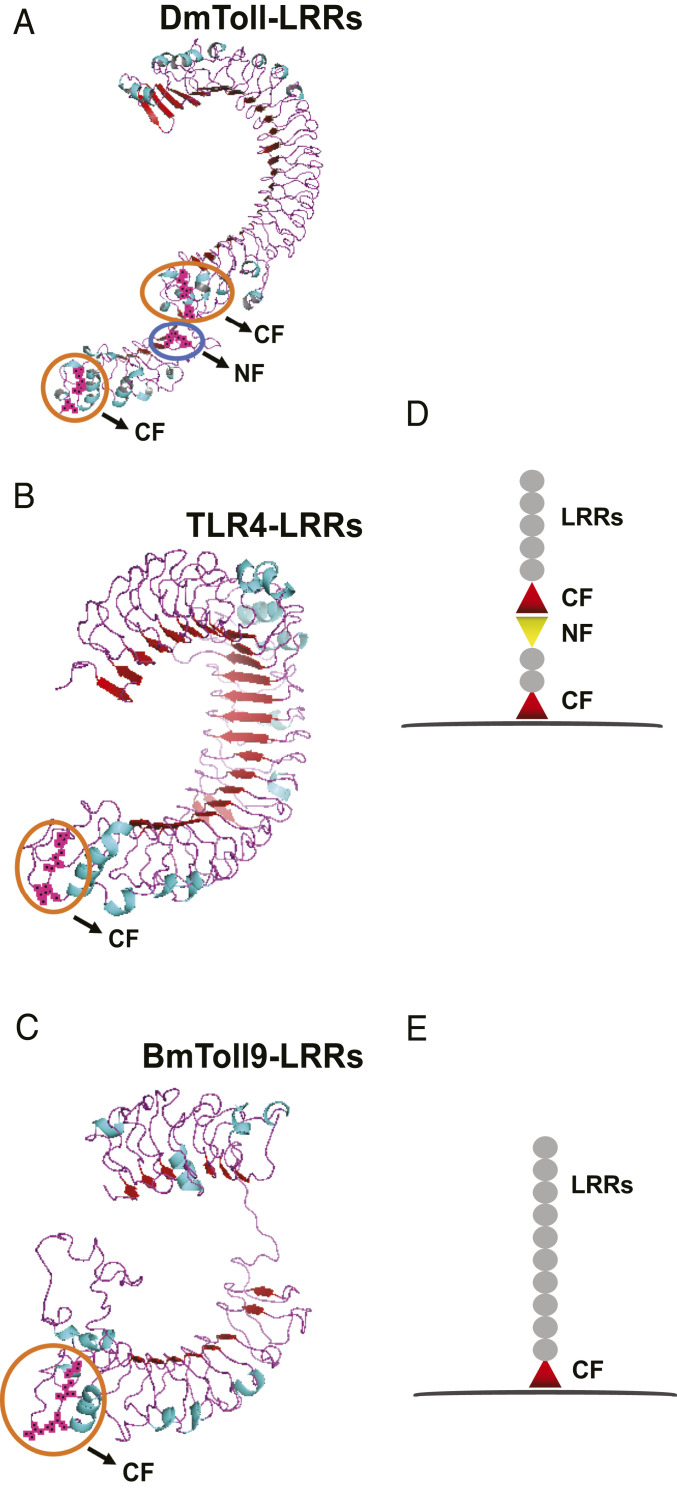
Insect Toll9 Evolutionarily Clusters with Mammalian TLR4.
Consistent with previous studies (15, 19), a broader phylogenetic analysis of Toll family members that included B. mori indicated all arthropod Toll9 members cluster most closely with mammalian TLR4 receptors, while all other arthropod Tolls cluster separate from vertebrate TLRs (SI Appendix, Fig. S1). Mammalian TLR ectodomains contain a single cysteine cluster C-terminal to the LRRs (designated the CF motif) that is juxtaposed to the plasma membrane to generate what are known as V-type TLRs, whereas the ectodomains of most insect Tolls contain multiple cysteine clusters to form P-type TLRs (30, 31). We used B. mori Toll9 (BmToll9), murine TLR4, and D. melanogaster Toll-1 (DmToll) to generate three-dimensional structures (Fig. 1 A–C), which showed that TLR4 and BmToll9 have one CF motif, while DmToll has two CF motifs plus a third cysteine cluster at the N terminus (designated the NF motif) (Fig. 1 D and E). Expanding to all B. mori Tolls indicated that only BmToll9 had one CF motif, while other family members had two CF motifs or two CF motifs and one NF motif (SI Appendix, Fig. S2). Thus, BmToll9 and other Toll9 members share features with mammalian TLR4s that differ from other arthropod Tolls.
Fig. 1.
Structural analysis of DmToll, BmToll9, and murine TLR4. (A–C) Model structures for the ectodomains of (A) Drosophila Toll (DmToll-1), (B) Mus musculus TLR4, and (C) B. mori Toll9 (BmToll9). Structures for DmToll-1 (PDB ID code 4LXR) and TLR4 (PDB ID code 3VQ1) were obtained from the PDB, while the model structure for BmToll9 was generated by SWISS-MODEL and PyMOL using human TLR4 (PDB ID code 3FXI) as the template. The cysteine clusters at the C terminus and N terminus of the LRRs are shown. (D and E) Schematic diagrams of the ectodomains from insect Tolls (D) and mammalian TLRs and insect Toll9 members (E).
BmToll9 Colocalizes with BmMD-2A and BmMD-2B on the Surface of B. mori Immune Tissues.
Focusing on B. mori larvae, we detected expression of BmToll9 in three tissues with immune functions (fat body, hemocytes, midgut) in the absence of immune challenge (SI Appendix, Fig. S3 A and B). Consistent with domain structure, we also detected BmToll9 on the surface of fat body cells, hemocytes, and midgut cells (SI Appendix, Fig. S3C). As previously noted, mammalian TLR4s require the ML protein MD-2 for LPS recognition and downstream signaling (32–34), while B. mori encodes three ML proteins that, as previously noted, were named BmEsr16, BmPP, and BmML-1 (27) but in this study were called BmMD-2A, BmMD-2B, and BmML-1, respectively. Antibodies detected BmMD-2A, BmMD-2B, and BmML-1 in fat body, hemocyte, midgut, and cell-free hemolymph (plasma) samples, which strongly suggested each was secreted (SI Appendix, Fig. S3D). Immunocytochemical assays also indicated that BmMD-2A and BmMD-2B colocalized with BmToll9 on the surface of fat body cells, hemocytes, and midgut cells (SI Appendix, Fig. S4).
Neutralization Assays Implicate BmToll9 in LPS-Induced Expression of Antimicrobial Peptide Genes.
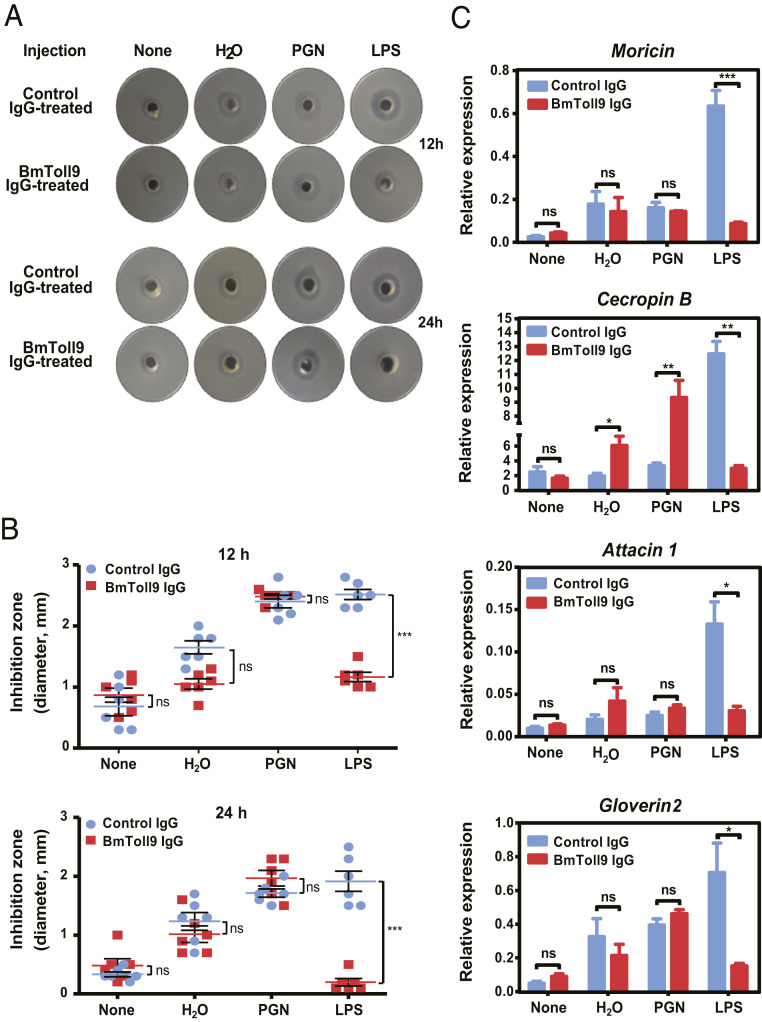
To assess whether BmToll9 is involved in LPS-induced signaling, we first conducted an in vivo neutralization experiment by injecting larvae with affinity-purified anti-BmToll9 or preimmune immunoglobulin G (IgG) (control) followed 1 h later by no second injection, a second injection of LPS-free H2O, or a second injection of H2O containing ultrapure PGN or LPS. Hemolymph and fat body were then collected 12 or 24 h later to assess antimicrobial activity. For larvae injected with preimmune IgG, inhibition zone assays with hemolymph showed that antimicrobial activity was higher in larvae that were challenged with PGN or LPS than H2O or no second injection (Fig. 2 A and B). For larvae injected with BmToll9 IgG, a similar trend was observed in response to PGN, H2O, or no second injection, whereas no increase in inhibition zone occurred when challenged with LPS (Fig. 2 A and B). In turn, LPS induced the expression of several AMP genes in the fat body including moricin, cecropin B, attacin 1, and gloverin 2, when larvae were first injected with preimmune (control) IgG, but expression was significantly lower when larvae were first injected with BmToll9 IgG (Fig. 2C). Expression of cecropin B but not the other AMP genes also increased when larvae were injected with water or PGN after first being injected with BmToll9 IgG (Fig. 2C). We were unclear why this occurred although it could reflect potential differences in cross-talk between pathways or competition between shared components that were differentially affected by first injecting larvae with BmToll9 IgG followed by water or PGN. Overall, though, the preceding results suggested a role for BmToll9 in LPS-induced expression of select AMP genes.
Fig. 2.
Anti-BmToll9 IgG blocks LPS-induced immune responses in B. mori larvae. Day 3 fifth-instar B. mori larvae were preinjected with purified anti-BmToll9ecto IgG or preimmune IgG (control), and then injected with ultrapure LPS-K12, PGN-K12, or H2O 1 h later. Larvae with no second injection served as a control (none). Hemolymph and fat body were collected 12 and 24 h after a second injection for antibacterial and qRT-PCR assays. (A) Representative inhibition zone assays using hemolymph and E. coli. (B) Mean diameter (mm) ± SEM of six inhibition zone assays. (C) Mean relative expression ± SEM of select antimicrobial peptide genes in fat body as determined by qRT-PCR. The expression of AMP genes in the fat body of larvae injected with preimmune IgG and no second injection (none) served as the reference for calculating the relative expression of each gene. Three biological replicates were generated for each treatment. Significant differences between the preimmune IgG and anti-BmToll9 IgG groups for each second injection treatment were determined by t test with significance indicated by asterisks: *P < 0.05, **P < 0.001, ***P < 0.0001; ns, not significantly different.
The BmToll9 Ectodomain Binds to BmMD-2A and BmMD-2B.
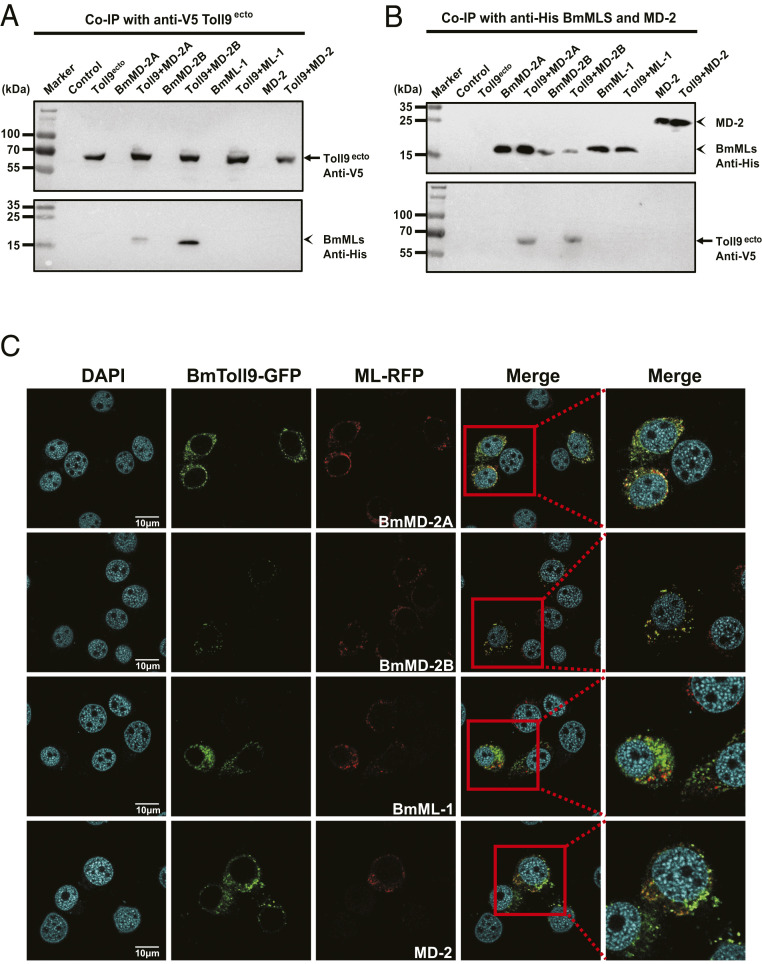
We next examined if ML proteins and BmToll9 directly interact by conducting coimmunoprecipitation (Co-IP) assays in an established B. mori cell line (BmN cells). RT-PCR and immunoblotting assays detected no expression of BmToll9 at the RNA or protein level in BmN cells but did detect expression and secretion of BmMD-2A, BmMD-2B, or BmML-1 (SI Appendix, Fig. S5 A and B). We therefore cloned BmToll9, its ectodomain (BmToll9ecto), and green fluorescent protein (GFP) (control) into pIEx-4/V5 for expression with V5 epitope tags while mouse MD-2 plus BmMD-2A, BmMD-2B, and BmML-1 were cloned into pIEx-4/His for expression with His epitope tags. Anti-V5 recognized recombinant BmToll9, BmToll9ecto, and GFP in transfected BmN cells but did not recognize any proteins in nontransfected cells (SI Appendix, Fig. S5C). As expected, we also detected BmToll9ecto but not BmToll9 in the medium (SI Appendix, Fig. S5C). Anti-His recognized recombinant MD-2, BmMD-2A, BmMD-2B, and BmML-1 in both transfected cells and conditioned medium but did not recognize any proteins in nontransfected cells including native BmMD-2A, BmMd-2B, or BmML-1 (SI Appendix, Fig. S5D). These results collectively indicated that conditioned medium containing recombinant BmToll9ecto and ML proteins could be used in Co-IP assays, which was done by mixing medium containing each combination of potentially interacting proteins with anti-V5 and anti-His antibodies. Results strongly supported that BmMD-2A and BmMD-2B bound to BmToll9ecto but BmML-1 and MD-2 did not (Fig. 3 A and B). We then generated a second set of expression constructs in which BmToll9 was tagged with GFP and each ML protein was tagged with red fluorescent protein (RFP). Cotransfection followed by confocal microscopy showed the colocalization of BmToll9 with BmMD-2A or BmMD-2B on the surface of BmN cells, whereas no colocalization was detected with BmML-1 or MD-2 (Fig. 3C).
Fig. 3.
BmToll9 binds and colocalizes with BmMD-2A and BmMD-2B. (A and B) Outcomes of Co-IP experiments. Culture medium from untransfected BmN cells (control), or BmN cells expressing BmToll9ecto–V5 (Toll9ecto), BmMD-2A–His (BmMD-2A), BmMD-2B–His (BmMD-2B), BmML-1–His (BmML-1), or murine MD-2–His (MD-2) was collected alone or mixed. (A) Adding an anti-V5 antibody plus protein G beads to each sample immunoprecipitated BmToll9ecto from culture medium containing Toll9ecto alone or when medium containing Toll9ecto was mixed with each ML protein (Toll9 + BmMD-2A, Toll9 + BmMD-2B, Toll9 + ML-1, and Toll9 + MD-2) as detected by probing the immunoblot (Upper) with anti-V5. Probing the immunoblot (Lower) with anti-His shows that only BmMD-2A and BmMD-2B were coimmunoprecipitated. (B) Reciprocal experiment of adding anti-His antibody plus protein G beads to each of the samples. Anti-His plus protein G beads immunoprecipitated each of the ML proteins in the samples as detected by probing the immunoblot (Upper) with anti-His. Probing the immunoblot (Lower) with anti-V5 shows that only BmMD-2A and BmMD-2B coimmunoprecipitated with BmToll9ecto. (C) Confocal microscopy images showing BmN cells coexpressing recombinant BmToll9–GFP (green) plus BmMD-2A–RFP, BmMD-2B–RFP, BmML-1–RFP, or murine MD-2–RFP (red). BmToll9 colocalized with BmMD-2A and BmMD-2B on the surface of BmN cells as indicated by yellow in the merged images but did not colocalize with BmML-1 or MD-2. Nuclei were stained with DAPI. Cells were imaged 48 h posttransfection. (Scale bars, 10 μm.)
LPS Stimulates the Expression of Select AMP Genes in BmN Cells that Express BmToll9.
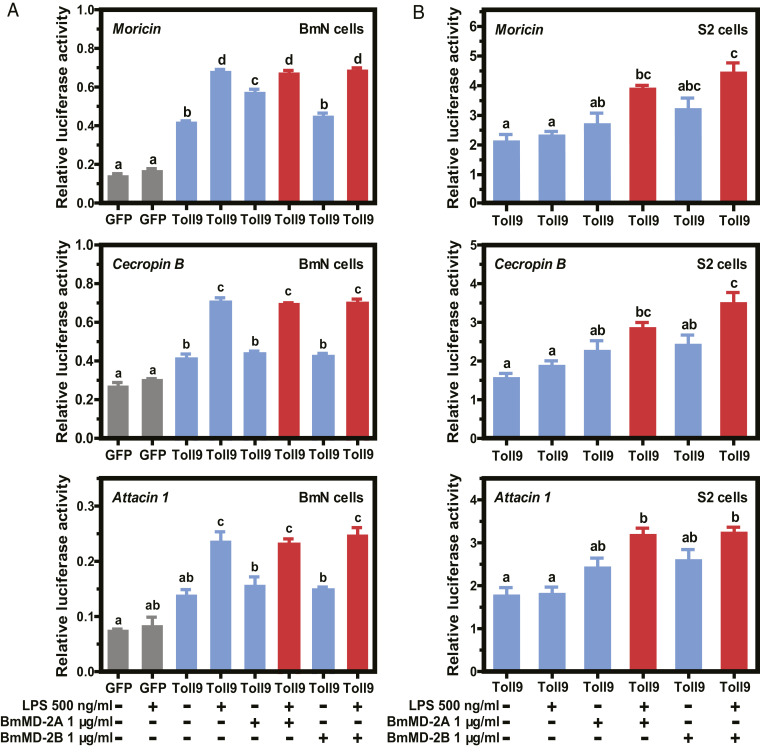
Given that our in vivo neutralization experiments implicated BmToll9 in LPS-induced expression of several AMP genes, we conducted dual-luciferase assays in BmN cells to determine if LPS activated the promoters of these genes when expressing BmToll9. This was approached by cotransfecting cells with pIEx-4-BmToll9 or pIEx-4-GFP (negative control) plus pGL3B constructs containing moricin-, cecropin B-, or attacin 1-luciferase reporters. Results showed that moricin, cecropin B, and attacin 1 promoter activity was lower in cells expressing GFP than BmToll9 (Fig. 4A). Adding LPS to the culture medium significantly increased reporter activity in cells expressing BmToll9, which was consistent with our in vivo data indicating that LPS also activates AMP gene expression in B. mori larvae (Fig. 4A). In contrast, adding LPS + BmMD-2A or LPS + BmMD-2B did not increase luciferase activity above LPS alone (Fig. 4A). This outcome could mean that BmToll9 does not require ML proteins for LPS recognition or downstream signaling, but we thought it more likely endogenous production of ML proteins in BmN cells (SI Appendix, Fig. S5B) resulted in recombinant ML proteins (SI Appendix, Fig. S5F) having no further effect when added to culture medium. We therefore transfected each pGL3B-luciferase reporter into Drosophila S2 cells that had been previously transfected to stably express BmToll9 (SI Appendix, Fig. S5E). Immunoblotting assays showed that BmToll9 was readily detected in stably transfected S2 cells (SI Appendix, Fig. S5E), whereas our antibodies to BmML proteins detected no cross-reacting products (SI Appendix, Fig. S5 B and F). Dual-luciferase assays in S2 cells expressing BmToll9 indicated that adding LPS alone to medium did not increase moricin, cecropin B, or attacin 1 promoter activity, whereas adding LPS + BmMD-2A or LPS + BmMD-2B increased activity which supported a role for two ML proteins from B. mori in LPS-induced signaling through BmToll9 (Fig. 4B). Since the effects of adding BmMD-2A or BmMD-2B to LPS similarly increased promoter activity, we also compared the effects of adding the same amount of BmMD-2A and BmMD-2B plus LPS on moricin promoter activity to assess whether these ML proteins potentially acted synergistically. However, results revealed no differences in moricin promoter activity when S2 cells were treated with LPS + BmMD-2A and BmMD-2B versus LPS + BmMD-2A or LPS + BmMD-2B (SI Appendix, Fig. S6). We thus concluded that BmMD-2A and BmMD-2B do not synergistically promote LPS-mediated expression of moricin through BmToll9.
Fig. 4.
BmMD-2A and BmMD-2B assist BmToll9 to activate LPS-induced AMP gene promoter activity in insect cell lines. Mean relative luciferase activity ± SEM in extracts prepared from cotransfected BmN (A) or S2 cells (B). BmN and S2 cells were cotransfected with pIEx-4–BmToll9–V5 or pIEx-4–GFP–V5 plus pGL3B-moricin, pGL3B-cecropin B, or pGL3B-attacin 1. Dual-luciferase assays were then conducted using extracts prepared from cells in which GFP was expressed with either no factors or LPS added for 12 h (gray bars); BmToll9 was expressed and either no factors or LPS, BmMD-2A–His, or BmMD-2B–His was added for 12 h (blue bars); or BmToll9 was expressed and LPS plus BmMD-2A–His or BmMD-2B–His were added for 12 h (red bars). Three biological replicates were generated per treatment. For each graph, different letters above bars indicate treatments significantly differed from one another (P < 0.05; ANOVA followed by a post hoc Tukey’s honestly significant difference [HSD] test).
The BmToll9 Ectodomain Can Partner with the TIR Domain of TLR4 to Transduce LPS-Induced Signaling.
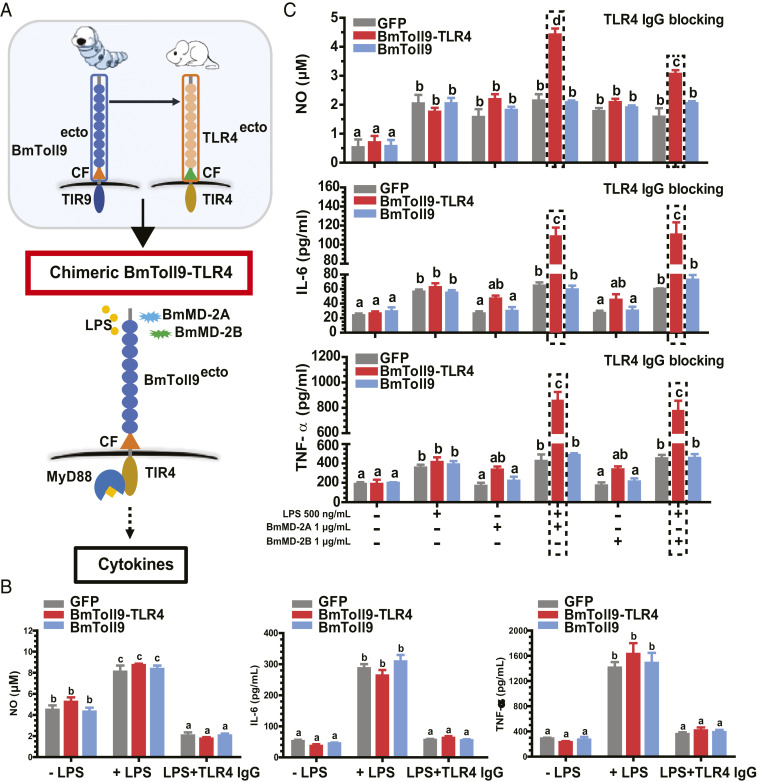
The murine macrophage-like RAW264.7 cell line is known to be LPS-responsive as measured by changes in morphology and effector molecule production due to endogenous expression of TLR4 and MD-2 (35). We therefore initially used this cell line to ask if BmToll9 or a chimeric receptor composed of the BmToll9 ectodomain plus mouse TLR4 transmembrane and TIR domains (BmToll9–TLR4) could confer signaling properties (Fig. 5A). We cloned BmToll9–TLR4, BmToll9, or GFP (negative control) into pcDNA3.1/HA for expression of each with a hemagglutinin (HA) epitope tag. Immunoblotting assays confirmed that transfected RAW264.7 cells expressed each construct while also remaining responsive to LPS as evidenced by a rapid change in morphology and increased production of nitric oxide (NO), IL-6, and TNF-α (Fig. 5B and SI Appendix, Fig. S7). However, adding a TLR4 antibody to the medium before LPS challenge greatly reduced NO, IL-6, and TNF-α production across each treatment, which suggested this response was primarily due to native TLR4 (Fig. 5B). We thus used this neutralization approach to assess whether BmToll9 or our BmToll9–TLR4 chimeric receptor could stimulate the production of these effector molecules if B. mori ML proteins were present. Adding BmMD-2A or BmMD-2B alone to medium containing anti-TLR4 had no effect on NO, IL-6, or TNF-α production across treatments (Fig. 5C). In contrast, adding LPS + BmMD-2A or LPS + BmMD-2B significantly increased NO, IL-6, or TNF-α production in cells expressing BmToll9–TLR4 but not BmToll9 or GFP (Fig. 5C).
Fig. 5.
LPS induces RAW264.7 cells expressing BmToll9–TLR4 to produce effector molecules if BmMD-2A or BmMD-2B is also present. (A) Schematic showing the BmToll9–TLR4 chimeric receptor, BmToll9, and TLR4, and assays conducted in RAW264.7 cells. (B) Mean titer ± SEM of NO, IL-6, and TNF-α produced by RAW264.7 cells expressing GFP–HA, BmToll9–HA, or BmToll9–TLR4–HA that were nonstimulated (−LPS), LPS-stimulated (+LPS), or pretreated with an anti-TLR4 antibody (2.5 μg/mL) and then LPS-stimulated. (C) Mean titer ± SEM of NO, IL-6, and TNF-α produced by RAW264.7 cells expressing GFP–HA, BmToll9–HA, or BmToll9–TLR4–HA that were pretreated with an anti-TLR4 antibody and then stimulated with LPS, BmMD-2A or BmMD-2B alone, or LPS plus BmMD-2A or BmMD-2B. For B and C, anti-TLR4 was added 6 h before the addition of LPS and/or BmML proteins followed by enzyme-linked immunosorbent assays 24 h poststimulation. For each graph, different letters above bars indicate treatments significantly differed from one another (P < 0.05; one-way ANOVA followed by a post hoc Tukey’s HSD test).
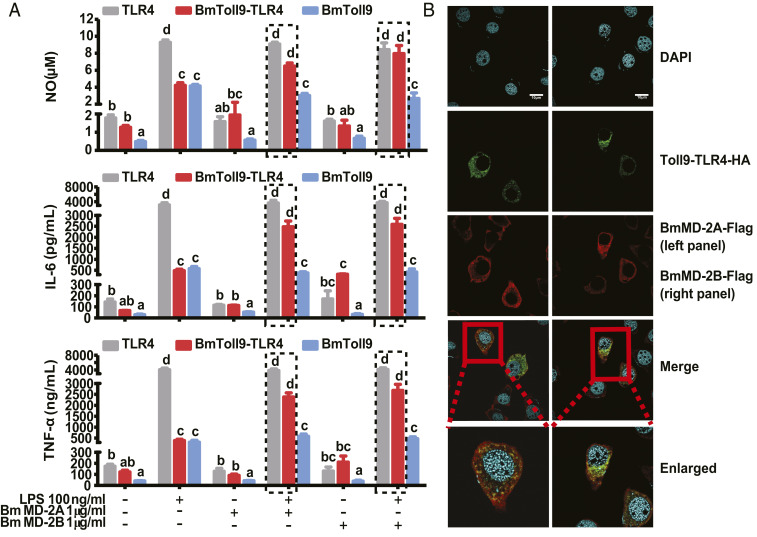
The preceding outcomes strongly suggested LPS can interact with B. mori ML proteins + BmToll9–TLR4 to stimulate downstream signaling in RAW264.7 cells. However, the presence of TLR4 remained a confounding effect even though activity was substantially neutralized by anti-TLR4. We therefore conducted a second set of assays using BMDM cells from TLR4 knockout (TLR4−/−) mice. Macrophage colony-stimulating factor (M-CSF) treatment induces BMDM cells to form CD11B/F480-positive cells (36) that exhibit a tentacled, adherent morphology (SI Appendix, Fig. S8 A and B). Cells were then transduced using adenoviruses carrying BmToll9–TLR4, BmToll9, or TLR4 with HA epitope tags. Immunoblotting assays confirmed that each recombinant protein was expressed in cells after transduction but as expected was not secreted into the medium (SI Appendix, Fig. S8C). Adding LPS strongly increased NO, IL-6, and TNF-α production in cells expressing TLR4–HA while modestly increasing production in cells expressing BmToll9–TLR4–HA or BmToll9–HA (Fig. 6A). In contrast, LPS + BmMD-2A or BmMD-2B did not increase the production of these effector molecules in cells expressing BmToll9–HA (Fig. 6A). However, cells expressing chimeric BmToll9–TLR4–HA produced similar amounts of NO, IL-6, and TNF-α as cells expressing TLR4–HA when treated with LPS + BmMD-2A or BmMD-2B (Fig. 6A). BMDM cells cotransduced to express BmToll9–TLR4–HA and BmMD-2A–Flag or BmMD-2B–Flag further showed that these proteins colocalized (Fig. 6B).
Fig. 6.
LPS induces activated BMDM cells expressing BmToll9–TLR4 to also produce effector molecules if BmMD-2A or BmMD-2B is present. (A) Mean titer ± SEM of NO, IL-6, and TNF-α produced by activated BMDM cells expressing TLR4–HA, BmToll9–HA, or BmToll9–TLR4–HA that were stimulated with LPS, BmMD-2A or BmMD-2B alone, or LPS plus BmMD-2A or BmMD-2B. For each graph, different letters above bars indicate treatments significantly differed from one another (P < 0.05; one-way ANOVA followed by a post hoc Tukey’s HSD test). (B) Confocal microscopy images showing that recombinant BmMD-2A–Flag or BmMD-2B–Flag colocalized with BmToll9–TLR4–HA on the surface of activated BMDM cells. Cells coexpressing BmMD-2A–Flag or BmMD-2B–Flag plus BmToll9–TLR4–HA were stimulated with LPS (100 ng/mL) for 12 h followed by detection of each protein using a mouse monoclonal HA antibody plus an Alexa Fluor 488 (green fluorescence) goat anti-mouse secondary antibody or a rabbit monoclonal Flag antibody plus Alexa Fluor 568 (red fluorescence) goat anti-rabbit secondary antibody. Nuclei were stained with DAPI. (Scale bars, 5 μm.)
Discussion
Toll/Toll-like receptors are known from four animal phyla (Nematoda, Arthropoda, Echinodermata, and Chordata), and while domain structure is overall conserved, structural and functional differences exist between taxa and receptor family members (15, 37). In the case of insects, bacterial PGN activates both the Toll and immune-deficiency (IMD) pathways in Drosophila to stimulate AMP gene expression while LPS does not (4, 23). As earlier noted, PGN likewise activates AMP gene expression in Lepidoptera (moths and butterflies) including B. mori and M. sexta (25, 26). However, prior studies indicated that LPS in association with BmMD-2A (BmEsr16) also stimulates the expression of select AMP genes (25–27). We thus hypothesized that BmMD-2A and possibly other ML proteins function as accessory factors that complex with one or more BmToll receptors in a manner similar to LPS inducing downstream signaling by TLR4–MD-2 in mammals (20, 38). However, no evidence previously existed that any insect Toll family members are LPS-responsive or interact with ML proteins.
In the first part of this study, our phylogenetic analysis of full-length arthropod Toll and mammalian TLR family members builds on earlier findings (15, 19) by showing that most insect Tolls cluster separate from mammalian TLRs, which suggests different evolutionary histories as well as potentially different functional interactions with ligands or intracellular signaling pathway components. In contrast, that insect Toll9 family members associate with the mammalian TLR branch more generally and TLR4s in particular suggests a different evolutionary history that could also potentially include functioning as LPS-responsive PRRs.
Results from the second part of our study experimentally support that BmToll9 functions as an LPS-responsive PRR that interacts with ML proteins in a manner similar to mammalian TLR4. Specifically, our in vivo neutralization, Co-IP, and dual-luciferase assays support that 1) BmToll9 binds BmMD-2A and BmMD-2B, and 2) BmToll9–ML complex formation mediates LPS-induced expression of several AMP genes in B. mori. Our results further indicate that LPS in combination with BmMD-2A and BmMD-2B can also activate the expression of inflammatory mediators in murine cells through a chimeric BmToll9–TLR4 receptor. Homo- versus heterodimer formation as well as the nature of the LRRs in the ectodomain of TLRs are both known to affect ligand binding affinities (39). Our results showing that BmToll9 does not bind murine MD-2 suggest differences exist in the binding affinities of BmToll9 and murine TLR4 ectodomains for different ML proteins. The ability of BmToll9–TLR4 but not BmToll9 to stimulate murine cells to produce inflammatory molecules in response to LPS further suggests differences in the TIR domain of BmToll9 prevent it from interacting with downstream components of the TLR4 signaling pathway.
ML family proteins, including MD-1, MD-2, NPC2, and GM2A, are widely distributed in plants, animals, and fungi (40). There is only one MD-2 gene and one Npc2 gene in the mouse and human genomes with NPC2 playing a role in cholesterol transport and MD-2 complexing with TLR4 (38, 41). In contrast, most insects encode multiple ML genes with 8 being present in M. sexta and D. melanogaster and 13 being present in Anopheles gambiae. However, insect ML proteins have to date also been somewhat arbitrarily named MD-2–like, NPC2-like, or ecdysone-responsive (Esr16-like) (42) without experimental evidence that particular family members have analogous functions to mammalian homologs. Results reported in this study provide evidence that BmMD-2A and BmMD-2B both complex with BmToll9. Our previous results further indicate that BmMD-2A more strongly binds to LPS than PGN or lipoteichoic acid while showing no binding to laminarin or mannan (27). Results from this study also support that BmMD-2B binds LPS while previous results indicate it may additionally bind certain glucans (43, 44). Thus, our results indicate that BmMD-2A and BmMD-2B play important roles in BmToll9 functioning as a PRR that binds LPS and activates the expression of several AMP genes. However, since ML proteins from B. mori (27, 43, 44) and other insects (45, 46) also bind other ligands, it is also possible ML proteins alone or in combination with other components could allow for combinatorial variation that has roles in regulating defense responses to multiple factors. In contrast, Drosophila NPC2a and NPC2e both bind LPS and PGN, yet overexpression of NPC2a and NPC2e in S2 cells only modulates AMP gene expression after PGN but not LPS treatment (28). The responsiveness of Drosophila to PGN but not LPS could thus in part be due to differences in how PGN and LPS interact with ML proteins and DmToll9. It is also possible that Drosophila is more sensitive to PGN than to LPS due to expansion of the PGN recognition protein (PGRP) family to 13 genes, which encode at least 17 PGRP proteins (47) that play diverse roles in regulating both the Toll and IMD pathways (4, 48). Altogether, the results of this study are significant because they show that BmToll9 functions as a PRR through complex formation with ML proteins that exhibit parallels to mammalian TLR4 members but differ from other arthropod Tolls. Future comparative studies will reveal whether LPS regulates antibacterial defenses in other arthropods besides Lepidoptera versus PGN being the more important pathogen-associated molecular pattern as seen in Drosophila.
Materials and Methods
Insects and Cell Lines.
B. mori (Dazao) larvae were obtained from the Institute of Sericulture and Agricultural Products Processing, Guangdong Academy of Agricultural Sciences, and reared on mulberry leaves at 25 to 28 °C. B. mori BmN cells were provided by Sheng Li, School of Life Sciences, South China Normal University, and maintained at 28 °C in Grace’s insect medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco). Drosophila S2 cells were originally purchased from American Type Culture Collection (https://www.atcc.org/) and maintained at 28 °C in SIM SF medium (Sino Biological) supplemented with 5% FBS. The murine RAW264.7 cell line was obtained from the Institute of Human Viruses, Sun Yat-sen University, and maintained at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% FBS in a 5% CO2 incubator.
Phylogenetic Analysis of Toll and TLR Family Members.
The E-INS-I alignment algorithm in MAFFT (version 7.407), which optimizes multidomain sequences separated by variable regions, was used to align complete Toll and TLR amino acid sequences. The Bayesian inference criterion method in ProtTest (version 3.4.2) was used to test and select the optimal amino acid substitution model, while phylogenetic trees were constructed using ExaBayes (version 1.4.2). Four Metropolis-coupled chains were run 1 × 106 generations (sampled every 500 generations) for the optimal amino acid substitution model with gamma distribution; the frequency of empirical amino acid occurrences and the fixed amino acid substitution model (VT model) were used for analysis. The final phylogenetic tree was visually edited and color-annotated in FigTree (version 1.4.4). Three-dimensional model structures were constructed with the SWISS-MODEL and PyMOL programs using Drosophila Toll (Toll-1) (Protein Data Bank [PDB] ID code 4LXR) as a template for all B. mori Tolls but BmToll9, and Homo sapiens TLR4 (PDB ID code 3FXI) was used as a template for BmToll9.
Antibodies.
BmToll9ecto (residues 20 to 220), BmMD-2A (residues 17 to 145), BmMD-2B (residues 19 to 154), and BmML-1 (residues 19 to 154) were PCR-amplified using gene-specific primers (SI Appendix, Table S2) and cloned into H6pQE60 (49) for expression of recombinant proteins with His epitope tags in Escherichia coli BL21. Recombinant proteins were purified by nickel-affinity chromatography and used as antigens to produce polyclonal antibodies that were commercially generated in rabbits and affinity-purified (ABclonal Biotechnology). Other antibodies used in the study were purchased from commercial sources as follows: mouse monoclonal anti-V5 (Cell Signaling Technology; 80076), rabbit monoclonal anti-His (Cell Signaling Technology; 12698T), rabbit monoclonal anti-Flag (Cell Signaling Technology; 14793), mouse monoclonal anti-HA (Cell Signaling Technology; 2367S), and mouse monoclonal anti-tubulin (Beyotime; AT819) antibodies.
Neutralization Assays.
Anti-BmToll9 or preimmune rabbit IgG was injected into the hemocoel of day 3 fifth-instar naïve B. mori larvae (5 micrograms per larva). Larvae were then injected with LPS-free H2O, ultrapure PGN-K12, or LPS-K12 from E. coli K12 (InvivoGen), or without a second injection. Hemolymph and fat body were collected 12 and 24 h after the second injection followed by inhibition zone and qRT-PCR assays that measured transcript abundance for the B. mori AMP genes moricin, cecropin B, attacin 1, and gloverin 2 with specific primers (SI Appendix, Table S2) using previously described methods (27). For qPCR assays, fat body, midgut, and hemocytes were collected from day 3 fifth-instar B. mori larvae, and total RNAs were isolated from these tissues and hemocytes using TRIzol Reagent (TaKaRa). First-strand complementary DNA (cDNA) was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa). qPCR reactions were performed using SsoFast EvaGreen Supermix (Bio-Rad) in 20-µL reaction mixtures that were run using a CFX96 qPCR machine (Bio-Rad). Each reaction contained 2 µL cDNA template, 0.5 µL each primer (10 µM) (SI Appendix, Table S2), 10 µL SYBR Green Mix, and 7 µL of H2O, and PCR was performed with the following program: 95 °C for 5 min, then 35 cycles of 95 °C for 5 s and 60 °C for 30 s, followed by a dissociation curve. The ribosomal protein 49 gene was used for normalization of the cDNA templates. Data were then analyzed by the Ct method as previously described (50).
Expression Constructs.
For assays conducted in insect cells, BmToll9, BmToll9ecto, GFP, BmMD-2A (BmEsr16), BmMD-2B (BmPP), BmML-1, and murine MD-2 were PCR-amplified using gene-specific primers (SI Appendix, Table S2) and then cloned in-frame into pIEx-4/V5 and pIEx-4/His (Novagen) that use the baculovirus IE1 promoter and C-terminal V5 or His tags. BmToll9, BmMD-2A, and BmMD-2B were also cloned into pIEx-4/GFP or pIEx-4/RFP that C-terminally fused each product with GFP or RFP. Plasmids were propagated in E. coli K12D31 and then midiprepped using previously established protocols (18). The promoter sequences for the B. mori moricin (1,000 bp), cecropin B (1,500 bp), and attacin 1 (1,000 bp) genes were PCR-amplified using specific primers (SI Appendix, Table S2) and ligated into the pGL3-Basic vector (Promega) for cloning of putative regulatory sequences that can drive expression of a cDNA encoding modified firefly (Photinus pyralis) luciferase. pGL3B constructs were propagated in E. coli and similarly processed to obtain plasmids for use in dual-luciferase assays. For assays conducted in mammalian cell lines, a BmToll9–TLR4 chimeric receptor was produced. Briefly, the DNA sequences encoding the ectodomain of BmToll9 and the transmembrane and TIR domain of mouse TLR4 were amplified by PCR using specific primers (SI Appendix, Table S2), B. mori and mouse cDNAs as templates, respectively, and the two PCR products were used as templates to amplify cDNA encoding BmToll9–TLR4 by overlapping PCR. cDNA sequences encoding BmToll9–TLR4, BmToll9, and murine TLR4 were PCR-amplified using specific primers (SI Appendix, Table S2) and cloned in-frame into pcDNA3.1/HA (Thermo Fisher) that uses the cytomegalovirus enhancer–promoter and adds a C-terminal HA epitope tag. BmMD-2A and BmMD-2B were cloned into pcDNA3.1/Flag for expression with a C-terminal Flag tag. Adenovirus constructs containing BmToll9–TLR4–HA, BmToll9–HA, and TLR4–HA were produced by Hanheng Biological using a two-plasmid system: shuttle plasmid (pHBAd series) and backbone plasmid (pBHGlox [deIta] E1, 3Cre) (51). Briefly, the shuttle plasmids containing the target genes (pHBAd–BmToll9–TLR4–HA, pHBAd–BmToll9–HA, and pHBAd–TLR4–HA) were constructed, and then the shuttle plasmid and the skeleton plasmid were used to transfect HEK293A cells. After plaque isolation and purification, culture medium containing the recombinant adenovirus of interest was amplified by 3× reinfection of HEK293A cells followed by collection, purification, and concentration with final titer determined by a plaque-forming unit method. All expression constructs were Sanger-sequenced (Sangon Biotech) to confirm sequence identity. Viruses were aliquoted and stored at −80 °C until used to infect activated BMDM cells.
Recombinant BmMD-2A and BmMD-2B.
Recombinant BmMD-2A and BmMD-2B were produced by stably transfecting S2 cells with pIEx-4–BmMD-2A–His or pIEx-4–BmMD-2B–His using previously established protocols (18). In brief, cells were seeded overnight in SIM SF medium with 5% FBS to ∼70% confluence, and then cotransfected with pCoBlast (Invitrogen) plus pIEx-4–BmMD-2A–His or pIEx-4–BmMD-2B–His using FuGENE (Sigma-Aldrich) according to the manufacturer’s instructions. Cells were selected 48 h posttransfection by addition of 25 μg/mL blasticidin S hydrochloride (15205; Sigma-Aldrich). BmMD-2A–His and BmMD-2B–His in the cell-culture medium from stable lines were then purified by Ni-NTA agarose and used in bioassays after determining protein concentration.
Immunofluorescence Assays with B. mori Immune Tissues and Cell Lines.
B. mori fat body, hemocyte, and midgut samples were collected from day 3 fifth-instar larvae. Fat body was fixed in 4% paraformaldehyde at 4 °C overnight. Hemolymph was directly dropped on a glass slide and saturated N-phenylthiourea solution was added to prevent melanization for 30 min, the slide was rinsed with phosphate-buffered saline (PBS), and adherent hemocytes were fixed in 4% paraformaldehyde for 30 min. Fixed samples were washed three times with 1% PBT (100 µL Triton X-100 in 10 mL PBS), each for 5 min at 4 °C, and incubated with 3% bovine serum albumin (BSA) in PBS to block nonspecific binding. Midgut was fixed in 4% paraformaldehyde at 4 °C overnight and then paraffin-embedded and sectioned (Biossci Biotechnology). Sections were placed at room temperature for 10 min, in xylene for 30 min, and then sequentially in absolute ethanol and 95, 85, 75, and 50% ethanol, each for 5 min, and finally in double-distilled H2O for 5 min. The sections were placed in a box filled with citric acid antigen repair solution, and the antigen was repaired by heating the sections in a microwave oven with medium heat for 8 min twice and then medium low for 7 min. Finally, the cooled sections were washed in PBS (pH 7.4) gently on a shaker three times, each for 5 min. Primary antibodies to B. mori ML proteins (1:500 dilution) were added to the samples and incubated at 20 °C for 2 h, followed by addition of Alexa Fluor 568 goat anti-rabbit secondary antibody (Abcam) (1:200 dilution) and incubation at 20 °C for 2 h. Samples were washed three times with 5% PBT, each for 15 min, to remove unconjugated fluorescent secondary antibody, and fluorescein isothiocyanate-labeled rabbit polyclonal antibody to BmToll9 (1:500 dilution) was added and incubated at 4 °C overnight. DAPI (1:10,000 dilution) (Life Technologies) was used to stain nuclei. BmN cells were cotransfected with pIEx-4–BmToll9–GFP plus pIEx-4–BmMD-2A–RFP or pIEx-4–BmMD-2B–RFP for 48 h using FuGENE as described above, and the cells were fixed with 4% paraformaldehyde, permeabilized with 1% Triton X-100, and blocked with 5% BSA blocking solution, and nuclei were stained with DAPI. All images were acquired using a confocal microscope (Olympus Microsystems; FV3000).
Coimmunoprecipitation Assays.
BmN cells were transfected with pIEx-4–BmToll9ecto–V5, pIEx-4–BmMD-2A–His, pIEx-4–BmMD-2B–His, pIEx-4–BmML-1–His, or pIEx-4–MD-2–His using FuGENE as described above. After confirming secretion of each recombinant protein by immunoblotting, culture media containing recombinant BmToll9ecto–V5 and each ML protein were precleared with protein G Sepharose (50% slurry; 17-0618-01; GE Healthcare) prior to Co-IP assays (16, 18). The cell-culture medium containing BmToll9ecto–V5 was then mixed with culture medium containing each ML protein followed by addition of anti-V5 or anti-His antibody (5 μg/mL). Each mixture was incubated at 4 °C for 10 h with gentle rocking. Protein G Sepharose beads (40 μL of a 50% slurry) in lysis buffer were added to the protein/antibody mixture and incubated overnight at 4 °C with gentle rocking. The Sepharose beads containing immunoprecipitated proteins were collected after centrifugation, washed three times with lysis buffer, resuspended in 50 μL of 1× sodium dodecyl sulfate sample buffer, boiled at 100 °C for 5 min, and used for Western blot analysis. These experiments were repeated at least three times (three independent biological samples or three independent cell cultures) with blots shown representing the outcome obtained in each independent replicate.
Dual-Luciferase Assays.
The pGL3-Basic vectors and renilla luciferase reporter plasmid (as an internal standard) (pRL-Actin 5) were purchased from Promega. The promoter sequences of B. mori antibacterial peptide genes moricin (1,000 bp), cecropin B (1,500 bp), and attacin 1 (1,000 bp) were cloned by PCR using specific primers (SI Appendix, Table S2) and ligated into the pGL3-Basic (pGL3B) vector to construct pGL3B recombinant vectors. The constructed pGL3B-promoter and pRL-Actin 5 plasmids were transfected into BmN or S2 cells. Dual-luciferase assays were performed using established methods (16). In brief, 8 × 104 cells per well were added to 24-well culture plates and maintained overnight in complete growth medium. BmN cells were then washed with serum-free medium, and then transiently cotransfected with pIEx-4–BmToll9, pGL3B, pGL3B-moricin, pGL3B-cecropin B, or pGL3B-attacin 1 firefly luciferase reporter plasmid (250 ng) and renilla luciferase reporter plasmid (25 ng) (as an internal standard) (pRL-TK; Promega) with FuGENE. S2 cells stably expressing BmToll9 were similarly transfected with the above pGL3B plasmids. Serum-free medium was replaced with the complete growth medium, followed 36 h later by the addition of LPS with or without recombinant ML proteins produced in S2 cells (see above). Firefly luciferase and renilla luciferase activities were then measured using the Dual-Luciferase Reporter Assay System (Promega) using a GloMax Multi Microplate Luminometer (Promega). Relative luciferase activity (RLA) was obtained as the ratio of firefly luciferase activity to renilla luciferase activity. The RLA obtained from BmN and S2 cells cotransfected with empty pIEx-4 and pGL3B (empty reporter vector) plasmids was used as the calibrator.
RAW264.7 and BMDM Cell Assays.
RAW264.7 cells were transfected by washing three times with PBS (pH 7.4) and resuspending in 100 μL of resuspension buffer R, then 5 μg of pcDNA3.1–GFP–HA, pcDNA3.1–BmToll9–HA, or pcDNA3.1–BmToll9–TLR4–HA plasmid was added and mixed using the Neon Transfection System (Thermo Fisher Scientific). Cells were then electroporated (voltage: 950 V; width: 20 ms; pulse: 1 pulse), resuspended in DMEM (Gibco) supplemented with 10% FBS, and seeded in a 24-well plate (8 × 105 cells per well in 0.5 mL medium). After 12 h, cell-culture medium was replaced with fresh medium, and cells were cultured an additional 36 h and then treated with LPS for 24 h or treated with monoclonal mouse anti-TLR4 antibody (BioLegend; 312804) (2.5 μg/mL) for 6 h followed by treating with LPS, BmML proteins, or LPS + BmML proteins for 24 h.
TLR4 knockout (TLR4−/−) homozygous mice (C57BL/10ScNJGpt, 6 to 8 wk old) were purchased from GemPharmatech. BMDM cells were isolated using previously described methods (52). Briefly, bone marrow was collected from femurs and maintained at 37 °C in DMEM supplemented with 10% FBS and M-CSF (50 ng/mL) (Neobioscience) in a 5% CO2 incubator. Cells were then seeded in 24-well plates (1 × 105 cells per milliliter) and cultured at 37 °C for 12 h in DMEM supplemented with 10% FBS and M-CSF (50 ng/mL) in a 5% CO2 incubator. Adenovirus carrying BmToll9–HA, TLR4–HA, or BmToll9–TLR4–HA (Hanheng Biotechnology) was added to BMDM cells at a multiplicity of infection of 100 for infection. After 2 h, cell-culture medium was replaced with fresh medium, cells were cultured an additional 12 h, and then treated with LPS, BmML proteins, or LPS + BmML proteins for 24 h.
Recombinant proteins expressed in RAW264.7 or BMDM cells were detected by immunoblotting using a mouse monoclonal anti-HA antibody. NO, IL-6, and TNF-α produced by RAW264.7 and BMDM cells in the cell-culture media were determined using NO (Promega), mouse IL-6 (Neobioscience), or TNF-α (Neobioscience) kits, respectively, following the manufacturers’ instructions.
BMDM cells were also transfected with pcDNA3.1–BmMD-2A–Flag or pcDNA3.1–BmMD-2B–Flag using a Neon Transfection System for 24 h, infected with adenoviruses carrying BmToll9–TLR4–HA for 24 h, and induced with LPS (100 ng/mL) for 12 h. BmToll9–TLR4–HA expressed in the BMDM cells was detected by anti-HA antibody (1:100) followed by Alexa Fluor 488 goat anti-mouse secondary antibody (1:200), while BmMD-2A–Flag or BmMD-2B–Flag was detected by anti-Flag antibody (1:100) followed by Alexa Fluor 568 (red fluorescence) goat anti-rabbit secondary antibody, and nuclei were stained with DAPI. Images were acquired using a confocal microscope (Olympus Microsystems; FV3000).
Data Analysis.
All assays were repeated at least three times using independently collected or prepared samples. Data were analyzed by Student’s t test or ANOVA followed by Tukey’s multiple-comparison tests using GraphPad.
Supplementary Material
Acknowledgments
This study was supported by the National Science Foundation of China (Grants 31970462 and 31970474) (to W.Y. and X.-Q.Y.), Natural Science Foundation of Guangdong Province (2018A0303130339) (to W.Y.), Guangdong Basic and Applied Basic Research Foundation (2020A1515010867) (to Y.Z.), and the US NSF (IOS1730276) and Pulliam Endowment (to M.R.S.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103021118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Anderson K.-V., Jürgens G., Nüsslein-Volhard C., Establishment of dorsal-ventral polarity in the Drosophila embryo: Genetic studies on the role of the Toll gene product. Cell 42, 779–789 (1985). [DOI] [PubMed] [Google Scholar]
- 2.Gay N.-J., Keith F.-J., Drosophila Toll and IL-1 receptor. Nature 351, 355–356 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto C., Hudson K.-L., Anderson K.-V., The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell 52, 269–279 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Lemaitre B., Hoffmann J., The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Lemaitre B., Nicolas E., Michaut L., Reichhart J.-M., Hoffmann J.-A., The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R., Preston-Hurlburt P., Janeway C. A. Jr, A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Shmueli A., Shalit T., Okun E., Shohat-Ophir G., The Toll pathway in the central nervous system of flies and mammals. Neuromolecular Med. 20, 419–436 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Akira S., Takeda K., Kaisho T., Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Takeda K., Evolution and integration of innate immune recognition systems: The Toll-like receptors. J. Endotoxin Res. 11, 51–55 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A., Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Hayashi F., et al., The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Hemmi H., et al., A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Hoshino K., et al., Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752 (1999). [PubMed] [Google Scholar]
- 14.Schwandner R., Dziarski R., Wesche H., Rothe M., Kirschning C.-J., Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274, 17406–17409 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Kanzok S.-M., et al., Origin of Toll-like receptor-mediated innate immunity. J. Mol. Evol. 58, 442–448 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury M., et al., Toll family members bind multiple Spätzle proteins and activate antimicrobial peptide gene expression in Drosophila. J. Biol. Chem. 294, 10172–10181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamoto M., et al., Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity 36, 658–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong X., Xu X.-X., Yi H.-Y., Lin C., Yu X.-Q., A Toll-Spätzle pathway in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 42, 514–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imler J.-L., Zheng L., Biology of Toll receptors: Lessons from insects and mammals. J. Leukoc. Biol. 75, 18–26 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Shimazu R., et al., MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189, 1777–1782 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West E.-E., et al., Pluripotent allospecific CD8+ effector T cells traffic to lung in murine obliterative airway disease. Am. J. Respir. Cell Mol. Biol. 34, 108–118 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitoh S., et al., Ligand-dependent Toll-like receptor 4 (TLR4)-oligomerization is directly linked with TLR4-signaling. J. Endotoxin Res. 10, 257–260 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Kaneko T., et al., Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Leulier F., et al., The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4, 478–484 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Rao X.-J., Yu X.-Q., Lipoteichoic acid and lipopolysaccharide can activate antimicrobial peptide expression in the tobacco hornworm Manduca sexta. Dev. Comp. Immunol. 34, 1119–1128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka H., et al., Lipopolysaccharide elicits expression of immune-related genes in the silkworm, Bombyx mori. Insect Mol. Biol. 18, 71–75 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Zhang R.-N., et al., An ML protein from the silkworm Bombyx mori may function as a key accessory protein for lipopolysaccharide signaling. Dev. Comp. Immunol. 88, 94–103 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Shi X.-Z., Zhong X., Yu X.-Q., Drosophila melanogaster NPC2 proteins bind bacterial cell wall components and may function in immune signal pathways. Insect Biochem. Mol. Biol. 42, 545–556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ao J.-Q., Ling E., Rao X.-J., Yu X.-Q., A novel ML protein from Manduca sexta may function as a key accessory protein for lipopolysaccharide signaling. Mol. Immunol. 45, 2772–2781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie L., Cai S.-Y., Shao J.-Z., Chen J., Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front. Immunol. 9, 1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leulier F., Lemaitre B., Toll-like receptors—Taking an evolutionary approach. Nat. Rev. Genet. 9, 165–178 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Park B.-S., et al., The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Miyake K., Endotoxin recognition molecules MD-2 and Toll-like receptor 4 as potential targets for therapeutic intervention of endotoxin shock. Curr. Drug Targets Inflamm. Allergy 3, 291–297 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Nagai Y., et al., Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 3, 667–672 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Chen Y.-F., et al., Zhankuic acid A isolated from Taiwanofungus camphoratus is a novel selective TLR4/MD-2 antagonist with anti-inflammatory properties. J. Immunol. 192, 2778–2786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno C., et al., Modulation of voltage-dependent and inward rectifier potassium channels by 15-epi-lipoxin-A4 in activated murine macrophages: Implications in innate immunity. J. Immunol. 191, 6136–6146 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Putnam N.-H., et al., Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Kim H.-M., et al., Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist eritoran. Cell 130, 906–917 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Paré A.-C., et al., An LRR receptor-teneurin system directs planar polarity at compartment boundaries. Dev. Cell 51, 208–221.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inohara N., Nuñez G., ML—A conserved domain involved in innate immunity and lipid metabolism. Trends Biochem. Sci. 27, 219–221 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Ko D.-C., Binkley J., Sidow A., Scott M.-P., The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc. Natl. Acad. Sci. U.S.A. 100, 2518–2525 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mészáros M., Morton D.-B., Comparison of the expression patterns of five developmentally regulated genes in Manduca sexta and their regulation by 20-hydroxyecdysone in vitro. J. Exp. Biol. 199, 1555–1561 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Miyake S., Yamano Y., Morishima I., Promoting protein, a silkworm hemolymph protein promoting in vitro replication of nucleopolyhedrovirus, binds to beta-glucans. Biosci. Biotechnol. Biochem. 69, 2012–2014 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Kanaya T., Kobayashi J., Purification and characterization of an insect haemolymph protein promoting in vitro replication of the Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 81, 1135–1141 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Dong Y., et al., Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2, e52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandiford S. L., et al., Cytoplasmic actin is an extracellular insect immune factor which is secreted upon immune challenge and mediates phagocytosis and direct killing of bacteria, and is a Plasmodium antagonist. PLoS Pathog. 11, e1004631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werner T., et al., A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 97, 13772–13777 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurata S., Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 42, 36–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee E., Linder M.-E., Gilman A.-G., Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol. 237, 146–164 (1994). [DOI] [PubMed] [Google Scholar]
- 50.Livak K.-J., Schmittgen T.-D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Mantwill K., et al., Inhibition of the multidrug-resistant phenotype by targeting YB-1 with a conditionally oncolytic adenovirus: Implications for combinatorial treatment regimen with chemotherapeutic agents. Cancer Res. 66, 7195–7202 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Marim F.-M., Silveira T.-N., Lima D. S. Jr, Zamboni D.-S., A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One 5, e15263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.