Figure 1. Inhibitors tested against HTLV-1 protease.
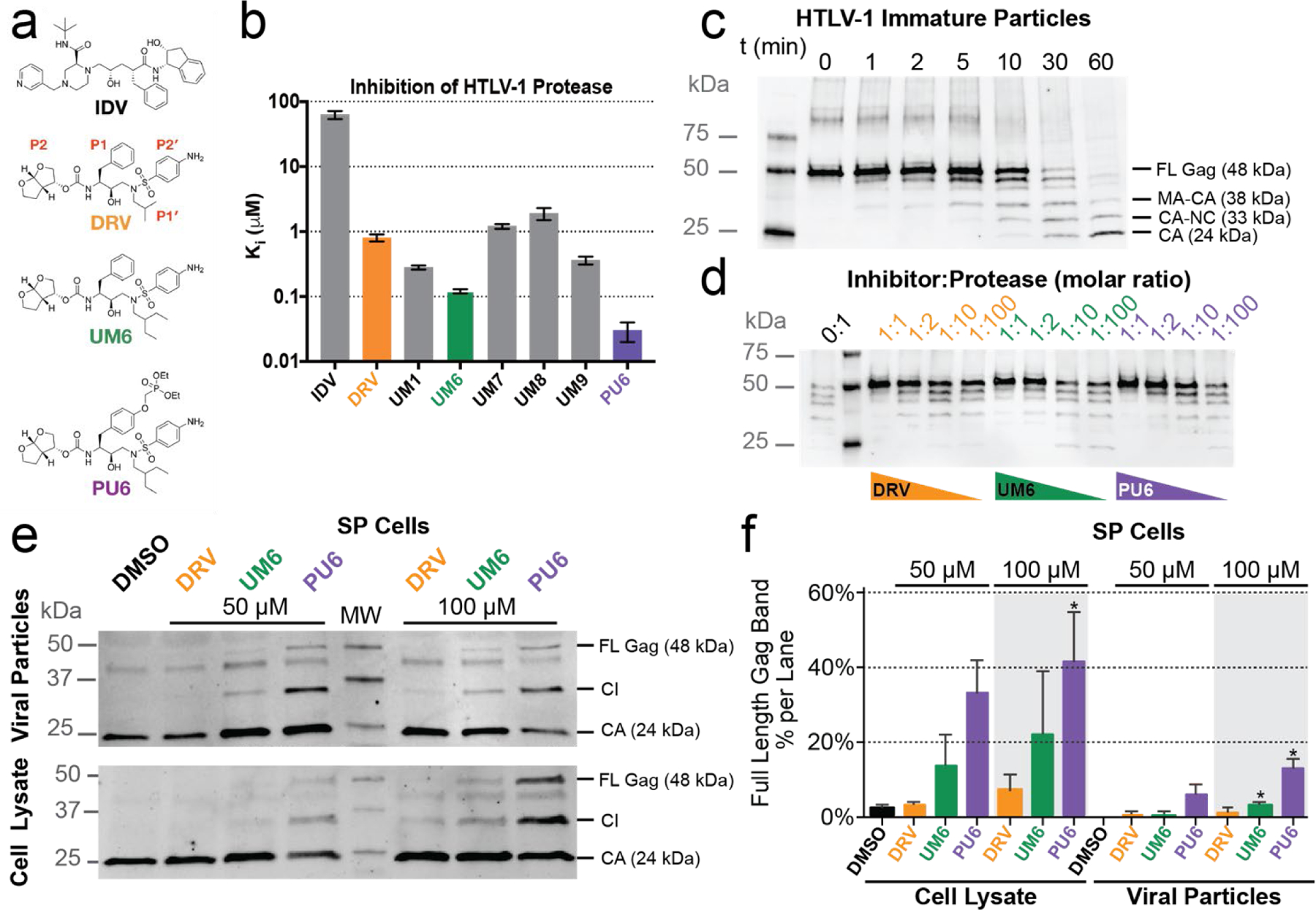
(a) Chemical structure of indinavir (IDV), darunavir (DRV) with P2–P2′ moieties labeled, and DRV analogs that exhibited improved potency. (b) Enzyme inhibition constants (Ki) for IDV, DRV, and DRV analogs against HTLV-1 protease. (c) HTLV-1 Gag cleavage by protease in immature virus particles. Gag cleavage by HTLV-1 protease (1 µM) at 37° C without inhibitor as a function of time. Cleavage products visualized with anti-HTLV-1 p24 (capsid). Expected cleavage products of full-length (FL) Gag are matrix-capsid (MA-CA), capsid-nucleocapsid (CA-NC), capsid (CA). (d) HTLV-1 protease inhibited in vitro against VLP Gag. Gag cleavage after 60 min with decreasing molar ratio of inhibitor to protease. At a 1:1 ratio (1 µM), no Gag cleavage products are observed for any inhibitor, while below 1:10 ratio (100 nM inhibitor) some p24 capsid is observed after 60 min. (e) HTLV-1 Gag cleavage from SP cell lysates and from particles released into cell culture supernatants were treated with 50 µM or 100 µM inhibitor for 48 hrs. Samples were then visualized by using an anti-HTLV-1 p24 antibody. (f) Immunoblot quantification of the FL Gag band as a percentage of total bands in each lane for the different SP cell drug treatments reported as the mean ± SEM (n=3). Band labels: full-length Gag (FL Gag), cleavage intermediates (CI), and capsid (CA). * denotes p-value < 0.05 with values compared to the DMSO control sample.
