Figure 1.
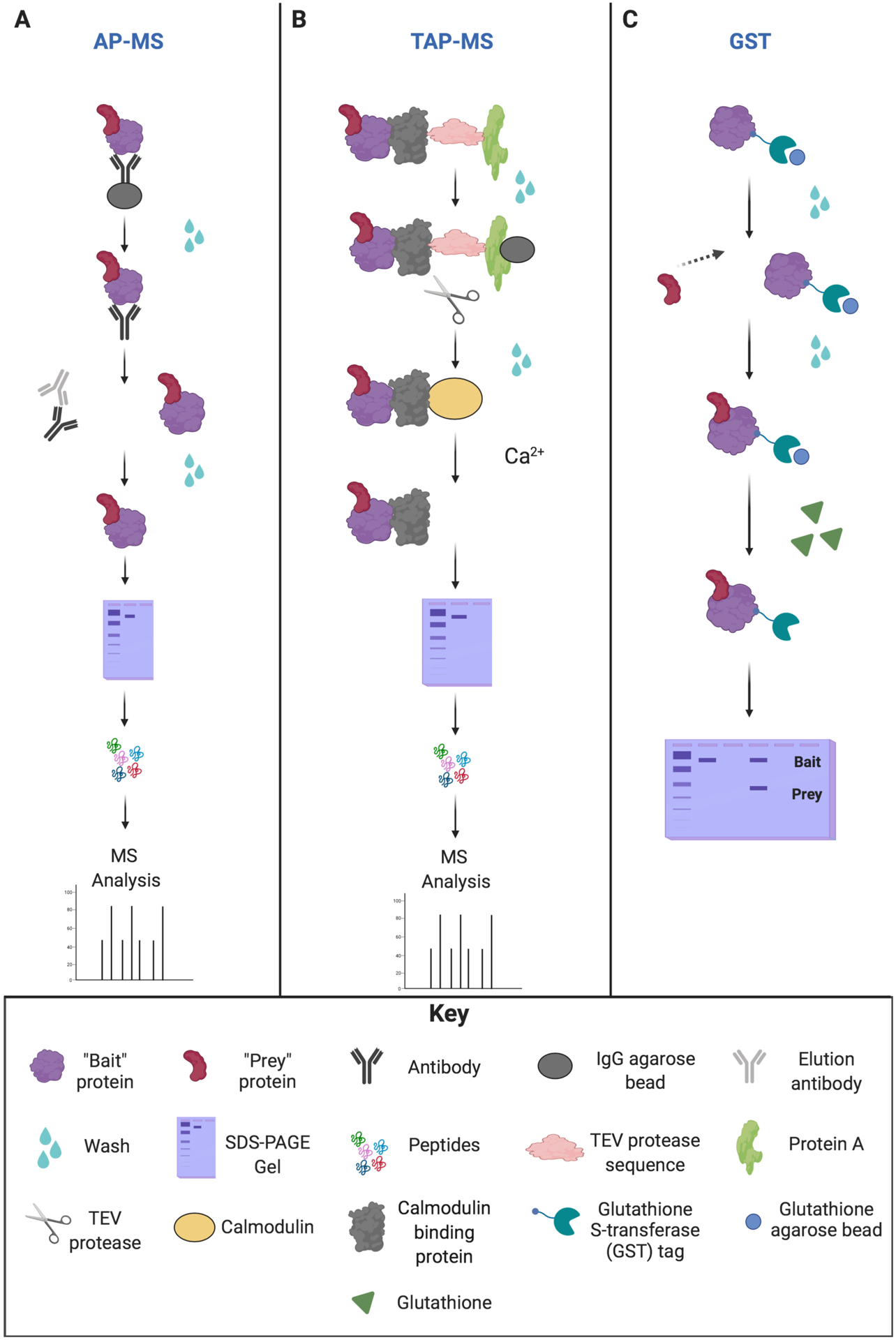
Overview of the different affinity pull-down approaches. (A) Affinity purification mass spectrometry (AP-MS) approach. AP-MS involves immobilization of “bait” protein on beads/resins, and then unbound proteins are washed away. The protein complexes are then eluted from the beads via antibody incubation. Protein complexes are purified and then separated by SDS-PAGE. Separated proteins are digested into small peptides, and the bait-prey interaction is analyzed by LC-MS/MS. (B) Tandem affinity purification (TAP-MS) approach. TAP-MS employs two different affinity purification steps for a protein complex containing the TAP-tagged protein. The first step immobilizes “bait” protein on an IgG bead/resin for protein A. Any unbound proteins are washed away. The protein complex is then immobilized on calmodulin-containing resins for the second affinity purification for calmodulin-binding protein (CBP). The addition of the tobacco etch virus (TEV) protease induces cleavage at the TEV recognition site. The addition of calcium (Ca2+) elutes the bound protein complex from the CBP. Protein complexes are purified and then separated by SDS-PAGE. Separated proteins are subsequently digested into small peptides that can be identified by LC-MS/MS. (C) Glutathione S-transferase (GST) pull-down assay. The recombinant fusion protein containing both GST and the “bait” is fixed to beads, and unbound proteins are washed away. The “prey” protein is added to the GST-tagged “bait” protein on beads to bind, and unbound “prey” proteins are washed away. Next, the bound protein complex is incubated with free glutathione, allowing for protein elution from the beads. Protein complexes are denatured by heating under reducing conditions. SDS-PAGE is then performed to separate the proteins, and the bait-prey interaction is analyzed by western blot.
