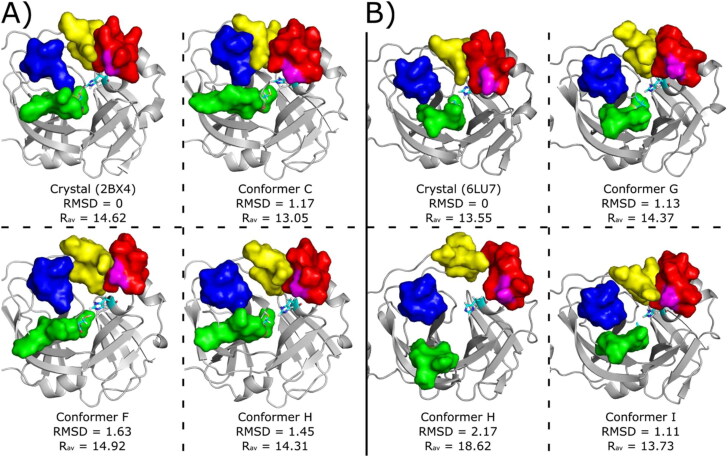
Figure 10.
Conformers used for ligand virtual screening and docking studies. The conformers were taken from the cluster centroids at the range between 0 and 10 kJ/mol in Figure 3. (A) MCoV1pro Crystallographic pose (PDB code 2BX4) and conformers C, F and H retrieved from metaDy simulations. (B) MCoV2pro crystallographic pose (PDB code 6LU7) and conformers G, H and I retrieved from metaDy simulations. For MCoV1pro only the conforme H is from well-tempered MetaDy, while for MCoV2pro only the conformer with this same label is from nontempered MetaDy. For all structures: RMSD (in Å) relative to crystal structure; Rav - average active site width (in Å); S11, S12, S2 and S4 loops are represented by surfaces coloured in blue, green, red and yellow, respectively. Catalytic residues His41 e Cys145 are represented in cyan sticks and residue Ala46 (MCoV1pro) or Ser46 (MCoV2pro) is represented in magenta surface. Animated gifs of both MetaDy for both Mpro can be seen in Supporting Information.