Figure 6 .
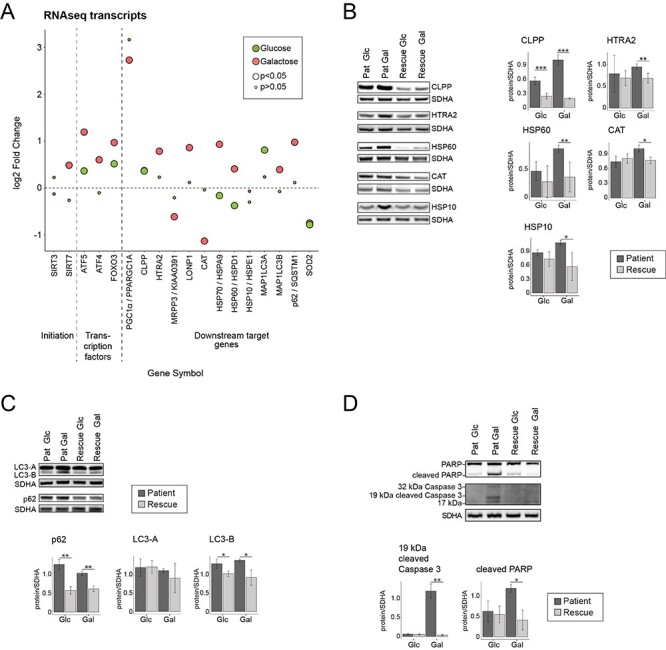
Metabolic dysfunction leads to an activation of the mitochondrial unfolded protein response (mtUPR) and increased autophagy. (A) Dot plot representing the fold change of the transcripts of mitochondrial integrated protein response. Fold change is represented in log2 scale. Large circles represent significant values (FDR < 0.05), small circles show non-significant values (FDR > 0.05). ‘Glucose’ (green) represents the comparison of patient versus rescue cells in glucose. ‘Galactose’ (red) represents the comparison of patient versus rescue cells in galactose. (B) Immunoblot analysis of the proteins involved in mitochondrial integrated stress response. Whole cell extracts from patient and rescue fibroblasts grown in glucose or galactose for 2 days were separated by SDS-PAGE and probed with antibodies against indicated proteins. SDHA was used as a loading control. Bar plot indicates the quantification of three (CLPP, HSP10, CAT), 4 (HTRA2, HSP60) immunoblot analyses, normalized to SDHA, where patient is depicted in dark grey and rescue in light grey, *P < 0.05, **P < 0.01, ***P < 0.001. (C) Immunoblot analysis of the proteins involved in autophagy. Whole cell extracts from patient and rescue fibroblasts grown in glucose or galactose for 2 days were separated by SDS-PAGE and probed with antibodies against indicated proteins. SDHA was used as a loading control. Bar plot indicates the quantification of three (LC3-A, LC3-B, p62) immunoblot analyses, normalized to SDHA, where patient is depicted in dark grey and rescue in light grey, *P < 0.05, **P < 0.01, ***P < 0.001. (D) Immunoblot analysis of the proteins involved in apoptosis. Whole cell extracts from patient and rescue fibroblasts grown in glucose or galactose for 2 days were separated by SDS-PAGE and probed with antibodies against indicated proteins. SDHA was used as a loading control. Bar plot indicates the quantification of three (cleaved PARP) and four (cleaved CC3) immunoblot analyses, normalized to SDHA, where patient is depicted in dark grey and rescue in light grey, *P < 0.05, **P < 0.01, ***P < 0.001.
