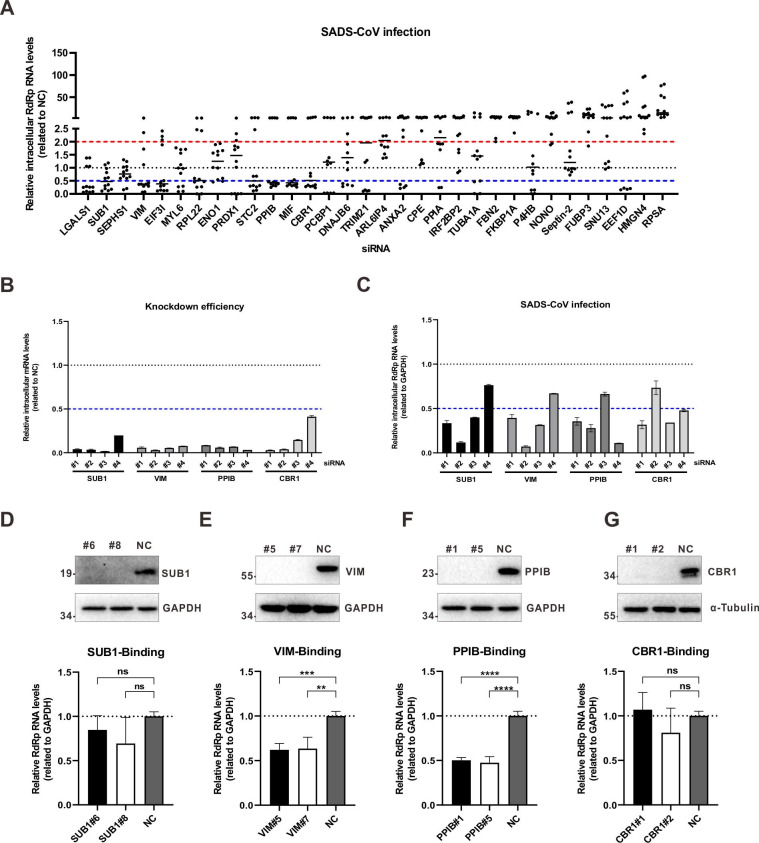
Fig. 4.
The effects of knockdown of S1-ΔNTD interactors on SADS-CoV infection. (A) For RNAi-based screening, three different siRNAs were designed for each gene. Each siRNAs were transfected into A549 cells. Forty-eight hours later, the cells were infected with SADS-CoV at an MOI of 0.01. Another 48 h later, intracellular mRNA was extracted. The relative SADS-CoV RNA level was measured using quantitative RT-PCR. Scramble siRNAs without target genes were used as negative controls (NCs). Data are representative of two independent experiments and three different siRNAs for each gene. (B, C) Four siRNAs were designed for SUB1, VIM, PPIB and CBR1 and used as (A). Relative levels of targeted genes (B) and the SADS-CoV RNA (C) were measured using quantitative RT-PCR. The data are representative of three independent experiments with similar results. Graphs show the means ± SD. (D, E, F, G) For CRISPR-Cas9-based knockout of the SUB1, VIM, PPIB and CBR1 genes, Cas9 and gene-specific sgRNAs were packaged into lentiviruses. A549 cells were transduced with lentivirus and selected under puromycin at least 1 week. Then, single cells were separated for expansion. Following Sanger sequencing and Western blotting (upper), the cell lines with frameshift mutations and merely protein expression were selected. For each gene, 2 cell lines were chosen to perform the virus-binding assay (below). SADS-CoV was incubated with A549 cells at an MOI of 0.1 at 4 °C for 1 h. Then, the cells were extensively washed with PBS to remove unbound viruses. Total RNA was extracted, and SADS-CoV genome level was measured with quantitative RT-PCR. A549 cells transduced with scramble sgRNA without target gene were used as negative control. The data are representative of two independent experiments with similar results. The graphs show means ± SD; n = 2. *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant, unpaired t-test.